Figure 3. DXO1 hydrolyzes the NAD moiety from NAD-RNA in vitro and affects in vivo NAD-RNA stability.
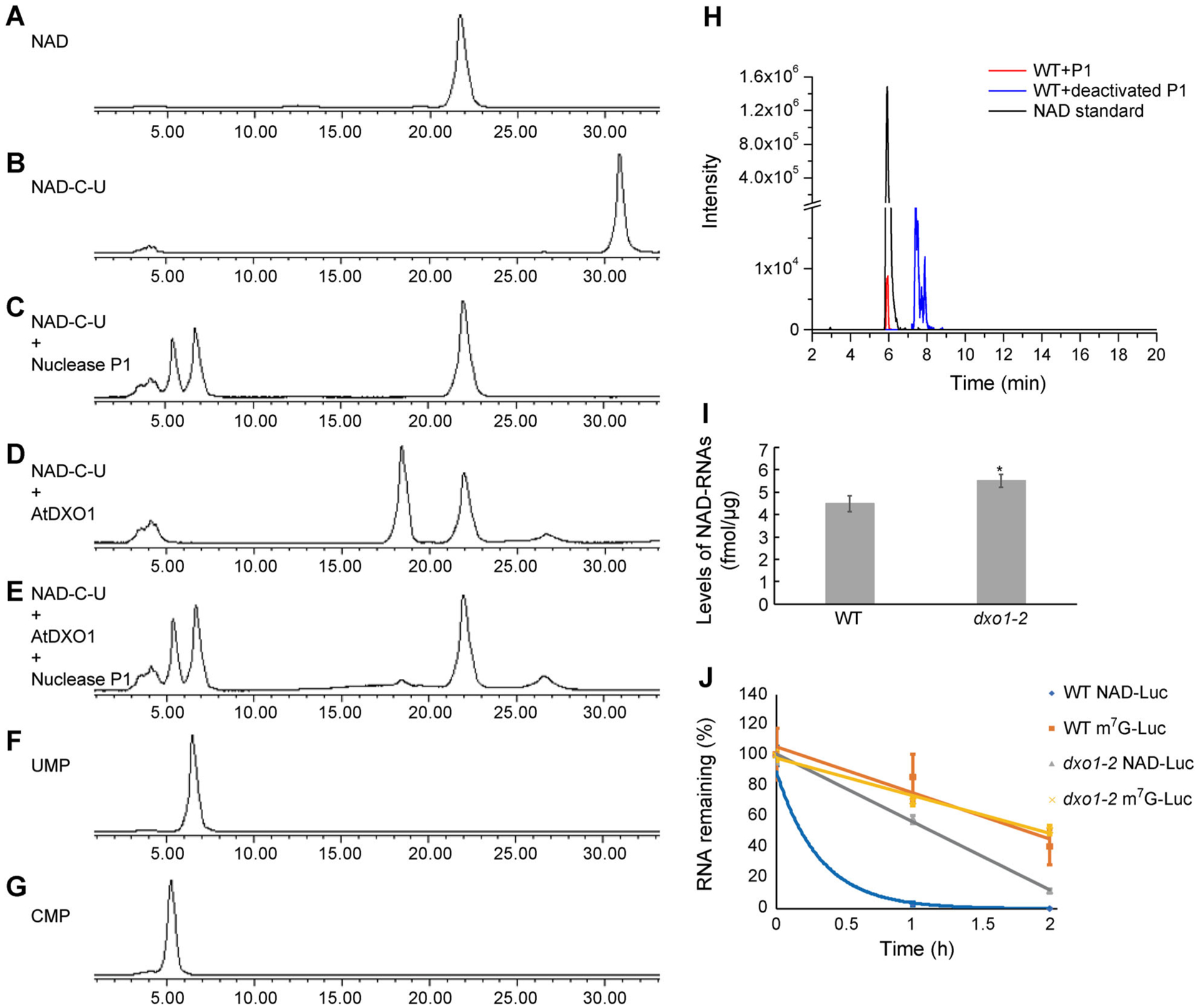
(A) and (B) HPLC peaks of NAD (A) and NAD-C-U (B). (C)–(E) HPLC peaks of NAD-C-U digested with 0.5 μM nuclease P1 at 37°C for 30 min (C), with 0.5 μM DXO1 at 37°C for 30 min (D), or with 0.5 μM DXO1 (30 min) followed by 0.5 μM nuclease P1 (30 min) at 37°C (E). (F) and (G) HPLC peaks of UMP (F) and CMP (G). (H) LC-MS detection of NAD from total RNAs of Arabidopsis after digestion by P1 nuclease. (I) Quantification of cellular NAD-capped RNA levels in WT and dxo1-2. Error bars represent standard deviation of three replicates; a t-test was used for statistical significance analysis; *Indicates statistically significant (P < 0.05). (J) Firefly luciferase mRNAs containing a 5′ NAD cap or an m7G cap and 3′ poly(A)60 were transfected into WT and dxo1-2 protoplasts. Protoplasts were harvested, and the remaining firefly luciferase mRNAs were detected by reverse transcription real-time PCR at the indicated time points. Error bars represent standard deviation of three replicates.
