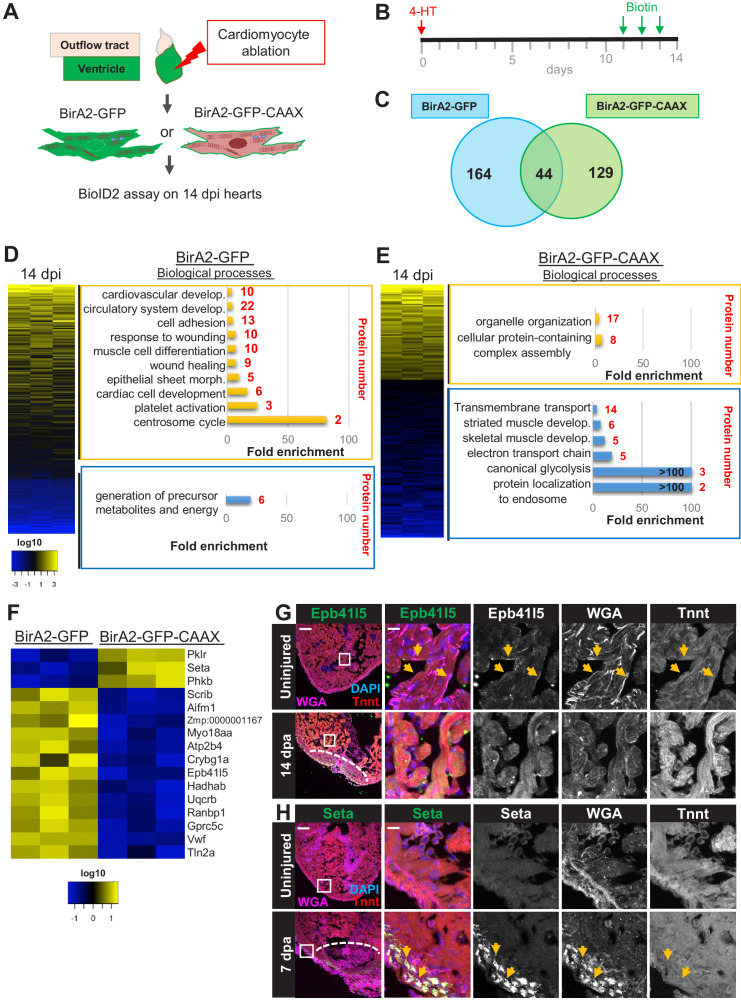
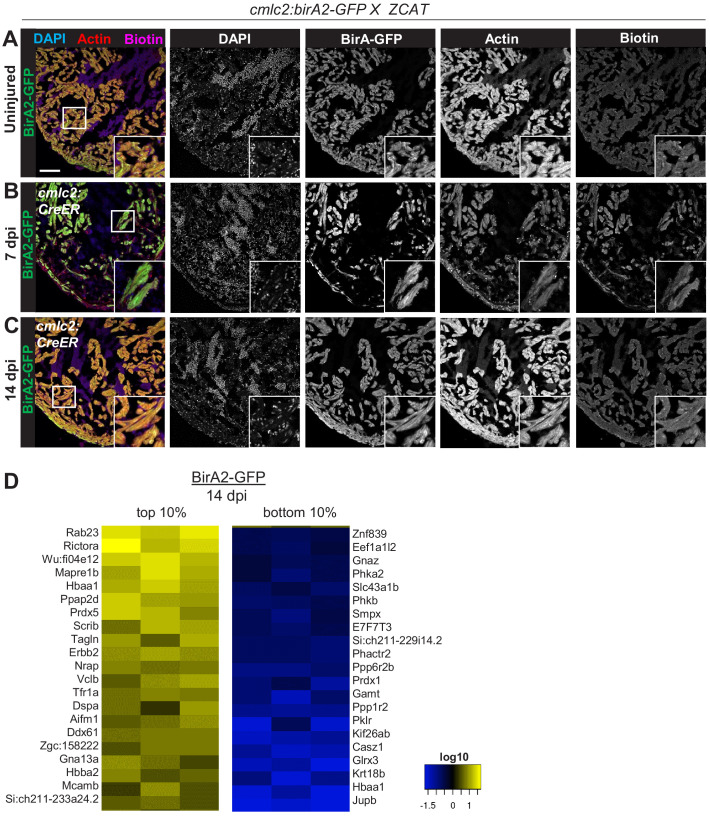
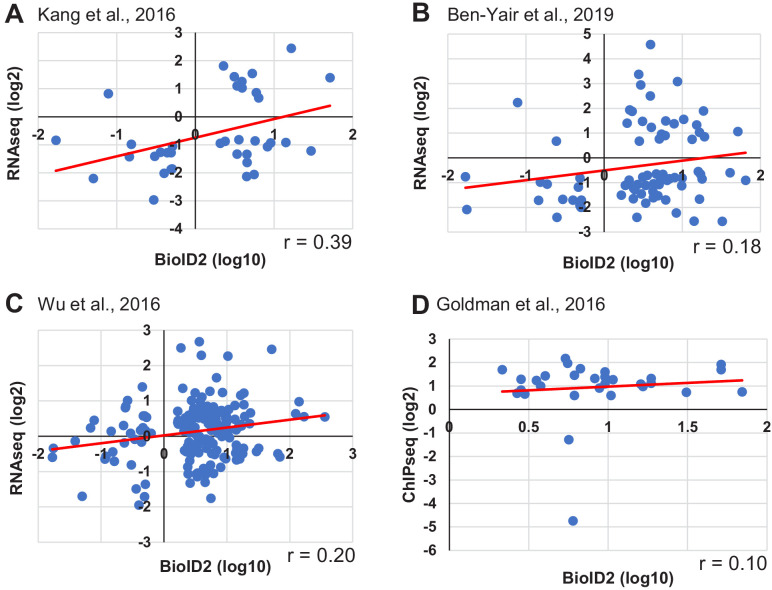
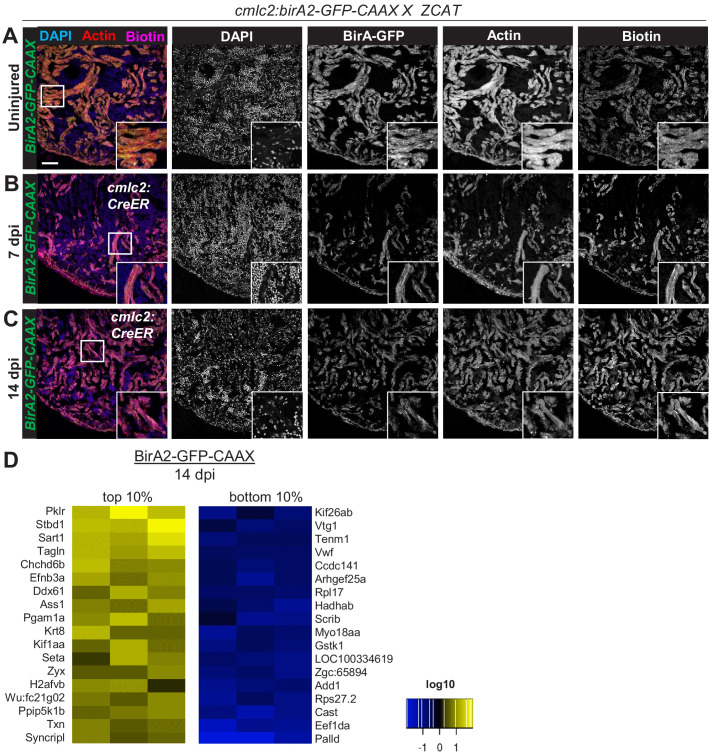
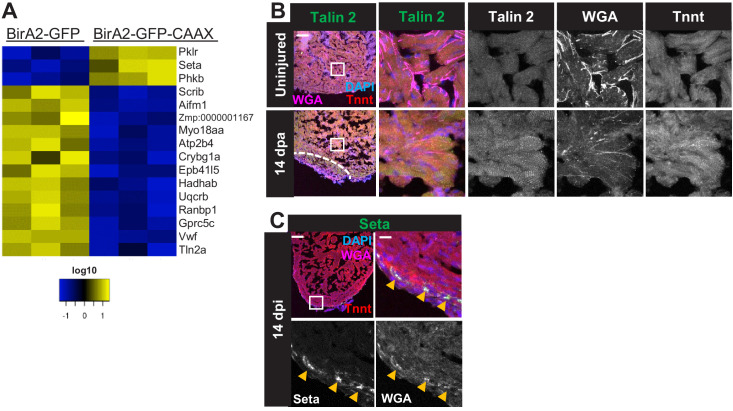
Figure 2. BioID2 identifies changes in membrane protein levels during heart regeneration.
(A) Schematic overview of experimental workflow. cmlc2:birA2-GFP or cmlc2:bira2-GFP-CAAX ventricles were collected as uninjured samples or 14 days after induced cardiomyocyte ablation (dpi). (B) Timeline of injury and biotin injections. (C) Venn diagram comparing proteins captured by either the whole-cell BioID2 assay or the membrane BioID2 data set that display at least a 1.5-fold change. (D) Summary of BioID2 analysis of cmlc2:birA2-GFP hearts during regeneration. Left: heat map of proteins found in triplicates of quantitative mass spectrometry analysis. 208 proteins displayed a 1.5-fold change during regeneration when compared to uninjured hearts (p<0.05). Of these, most protein levels increase during heart regeneration. Protein level changes in log10 scale. Right: gene ontology analysis of BirA2-GFP BioID2 data set. Over-representation test of biological processes for at least 1.5-fold increased (yellow) and at least 1.5-fold decreased proteins (blue). Fold enrichment is shown, and protein number is indicated in red. p<0.001, FDR < 0.04%. (E) Summary of BioID2 on cmlc2:bira2-GFP-CAAX hearts. Left: heat map of proteins found in triplicates of quantitative mass spectrometry analysis. 173 proteins displayed an at least 1.5-fold change when compared to uninjured hearts (p<0.05). Of these, most proteins decreased at the membrane during heart regeneration. Right: gene ontology analysis of BirA2-GFP-CAAX BioID2 data set. p<0.001, FDR < 0.05%. (F) Heat map of proteins that have been identified in the BioID2 whole-cell and membrane data sets and that display opposing changes in levels. Those shown changed at least 1.5-fold, p<0.05. Heat map summarizes fold changes measured in three separate pooled samples. (G) Immunofluorescence against indicated proteins in ventricles. Epb41l5 is localized to the plasma membrane (marked by wheat germ agglutinin [WGA] staining) in uninjured hearts, with cytoplasmic fluorescence signals increasing during regeneration of resected tissue (14 days post amputation [dpa]). (H) Seta is poorly detected in uninjured hearts, rising 7 days after resection injury in the epicardium and compact layer of the heart. Seta localizes around the nucleus and colocalizes with the membrane marker WGA. Scale bar in images, 50 μm; magnification scale bar: 10 μm.