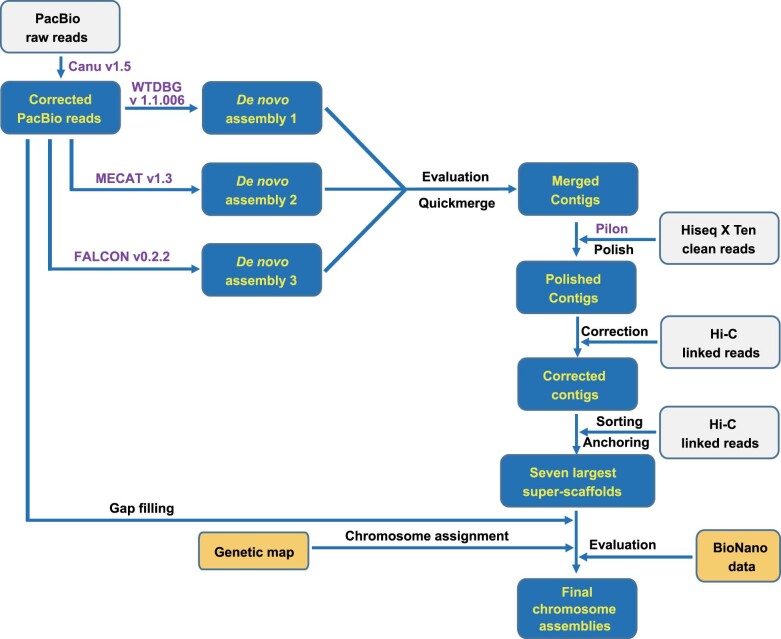
Extended Data Fig. 1. Pipeline for constructing the seven chromosome assemblies of Weining rye.
Multiple sequencing technologies and assembly strategies were applied to construct the genome sequence of Weining rye (Supplementary Tables 1–6). First, 497.01 Gb of long reads (62× genome coverage) generated by single molecule real-time sequencing (Pacbio Sequel), together with 430 Gb Illumina paired-end clean reads (54× genome coverage), were jointly used to assemble the genome into 63,244 contigs with an N50 size of 480.35 kilobases (kb). Second, six high-throughput chromatin conformation capture (Hi-C) libraries were constructed and yielded 1.87 billion clean paired-end reads, over 90% of which could be mapped to the initial assembly. This three-dimensional proximity information allowed us to correct, order, orientate and anchor PacBio contigs into super-scaffolds, with the seven largest ones selected as candidates of the seven chromosomes of Weining rye (Extended Data Fig. 2 and Supplementary Table 5), which were then subjected to gap filling with corrected PacBio reads and manual adjustment. Third, a genetic map (Extended Data Fig. 3), developed using 295 F2 individuals derived from crossing Weining rye with another rye variety (Jingzhou) (Supplementary Data 1), was used to assign the seven super-scaffolds onto chromosomes, which yielded the final 1 R to 7 R chromosome-scale assemblies. Approximately 96.02% of the assembly was covered with BioNano molecules (97× genome coverage), and a high consistency was found between the genome assembly of Weining rye and the BioNano genomic reads (Supplementary Fig. 1).