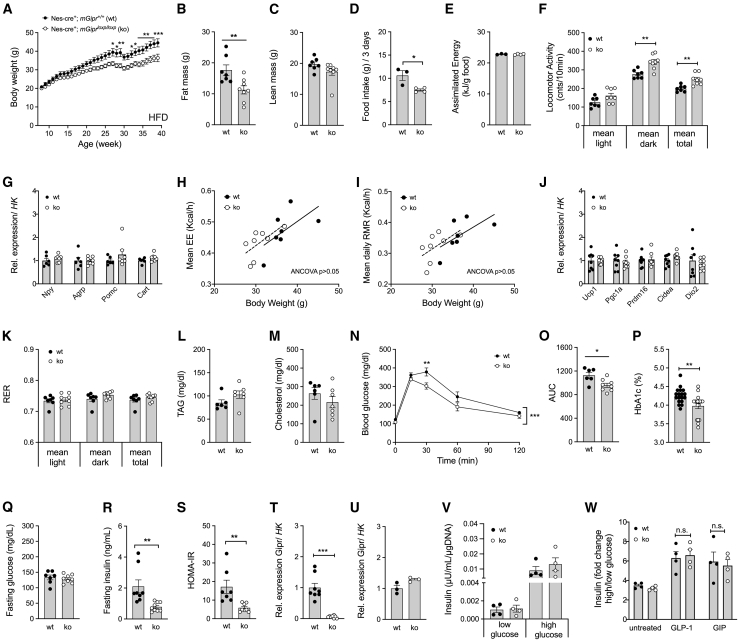
Figure 1.
Mice with CNS deletion of murine Gipr are protected from diet-induced obesity and glucose intolerance
(A–E) Body weight (A), body composition at the age of 28 weeks (B and C), food intake (D), and assimilated energy (E) in 42-week-old male C57BL/6J WT and CNS-Gipr KO mice (N = 7–8 mice each group) fed with a high-fat diet (HFD). Food intake and assimilated energy were assessed per cage in double-housed mice.
(F–I) Locomotor activity (N = 7–8 mice each group) (F), hypothalamic expression of genes related to food intake (6–7 mice each group) (G), and total energy expenditure (H) and resting metabolic rate (I) in 29-week-old male mice (N = 7–8 mice each group).
(J) Expression of genes related to BAT thermogenesis in HFD-fed male mice (N = 8 each genotype).
(K) Respiratory exchange ratio (RER) in 29-week-old male mice (N = 7–8 mice each group).
(L–O) Plasma levels of triglycerides (L) and cholesterol (M) (N = 6–7 each group) and intraperitoneal glucose tolerance (N and O) (N = 6–8 mice each group) in 42-week-old male mice.
(P) HbA1c (N = 18 mice each group; p = 0.0033).
(Q and R) Fasting levels of blood glucose (Q) and insulin (R) as well as HOMA-IR (S) in 42-week-old male mice (N = 7–8 each group).
(T and U) Relative expression of Gipr (corrected to housekeeping gene peptidylprolyl isomerase B; Ppib) in the hypothalamus (N = 8 mice each genotype) (T) and in isolated islets from WT and CNS-Gipr KO mice (N = 3 each group) (U).
(V and W) Glucose-stimulated insulin secretion (GSIS) in isolated islets under conditions of low (2.8 mM) and high glucose (16.8 mM) (V) and GSIS of isolated islets treated with or without 10 nM of either acyl-GLP-1 or acyl-GIP (W) (N = 4 mice each group). y axis in (W) represents the ratio of secreted insulin stimulated with high glucose (16.8 mM) to low glucose (2.8 mM).
Data represent means ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Longitudinal data (A and N) were analyzed using two-way ANOVA with time and genotype as co-variables and Bonferroni post hoc analysis for individual time points. Bar graphs (B–G, J–M, and O–W) were analyzed using two-tailed, two-sided t test. Data in (H) and (I) were analyzed using ANCOVA with body weight as co-variate.