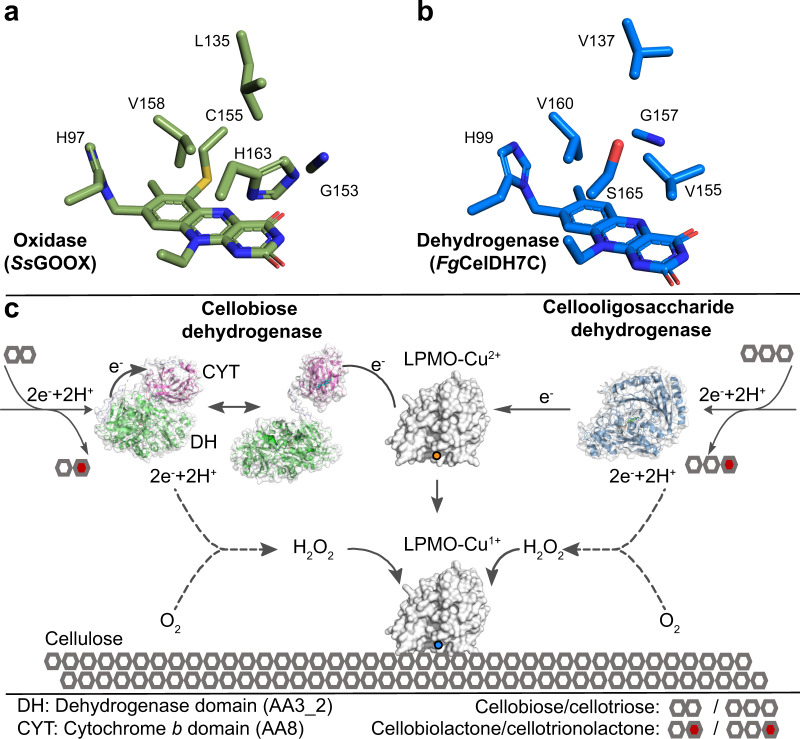
Fig. 6. Oxygen-binding cavity in AA7 oxidoreductases and a schematic model for AA7-LPMO interplay.
a, b Putative oxygen-binding cavity in typical AA7 oxidases and dehydrogenases, respectively. The substitution of conserved histidine and glycine in oxidases to serine (or other small residues) and valine in dehydrogenases, respectively, is observed. c Schematic model of the interplay of cellobiose dehydrogenase (CDH) versus the AA7 cellooligosaccharide dehydrogenase with LPMOs during cellulose degradation. Both dehydrogenase classes oxidise cellooligosaccharides to the corresponding lactones. The electrons harvested from this oxidation are stored in the FAD-cofactor and subsequently delivered directly to the LPMO in the case of AA7. The transfer of priming electrons in CDH proceeds first from the dehydrogenase domain to the cytochrome b haem domain in (closed form). A subsequent large conformational change to the open form is required to expose the haem domain to the LPMO active site for electron transfer. The dotted lines signify the low oxidase side-activity from both classes of dehydrogenases, which generates the H2O2 preferred co-substrate to fuel cellulose oxidative degradation by LPMOs. Low levels of H2O2 are also generated at the active site of free primed LPMOs, but this is left out from the figure for clarity.