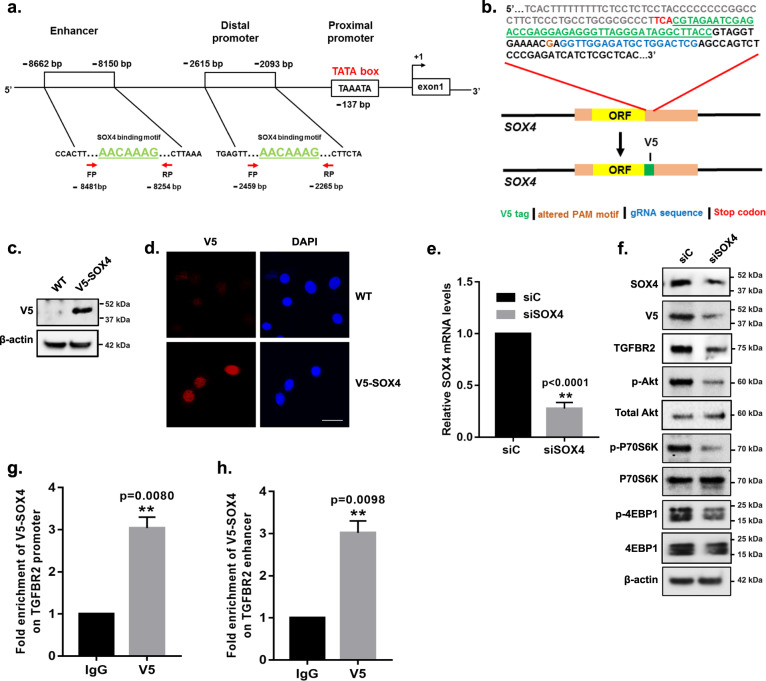
Fig. 4. SOX4 binds to regulatory sequences in TGFBR2 promoter and enhancer region.
a Schematic representation of the TGFBR2 regulatory regions upstream from the transcription start site. The proximal promoter with TATA box (red), the SOX4 binding motif, (AACAAAG) in the distal promoter and enhancer (green), and forward/reverse primer locations (red arrows) are noted. b Schematic of the CRISPR/Cas9 genome editing strategy used to insert a C-terminal V5 epitope tag (green) at the 3′ locus immediately upstream of the stop codon (red); the gRNA sequence (blue) and the altered PAM motif (brown) are highlighted. c Western blot and d immunofluorescence demonstrate V5 expression in the HCC1143 SOX4-V5 cell line; no expression is observed in the parental cell. The scale bar is 100 µm. e qRT-PCR demonstrates that SOX4 mRNA levels are significantly reduced by siSOX4 in HCC1143SOX4-V5 (p < 0.0001) relative to siControl transfected cells (50 nm, 96 h). f Western blot analyses demonstrated reduced protein expression of SOX4, V5, TGFBR2, and phosphorylated PI3K pathway marker proteins in HCC1143 SOX4-V5 cells transfected with siSOX4 relative to siControl-treated cells (96 h). g ChIP-qPCR using anti-V5 antibody demonstrated a 3.03-fold enrichment (p = 0.008, unpaired t-test) of V5-tagged Sox4 protein at the TGFBR2 promoter relative to IgG control. h A 3.01-fold enrichment of V5 protein (p = 0.01, unpaired t-test) was observed at the TGFBR2 enhancer in HCC1143SOX4-V5 cells relative to IgG control. Fold enrichment is normalized to PNOC promoter region, which is used as a negative control.