Figure 2. Eps15 and Fcho1 require multivalent interactions to assemble on membrane surfaces.
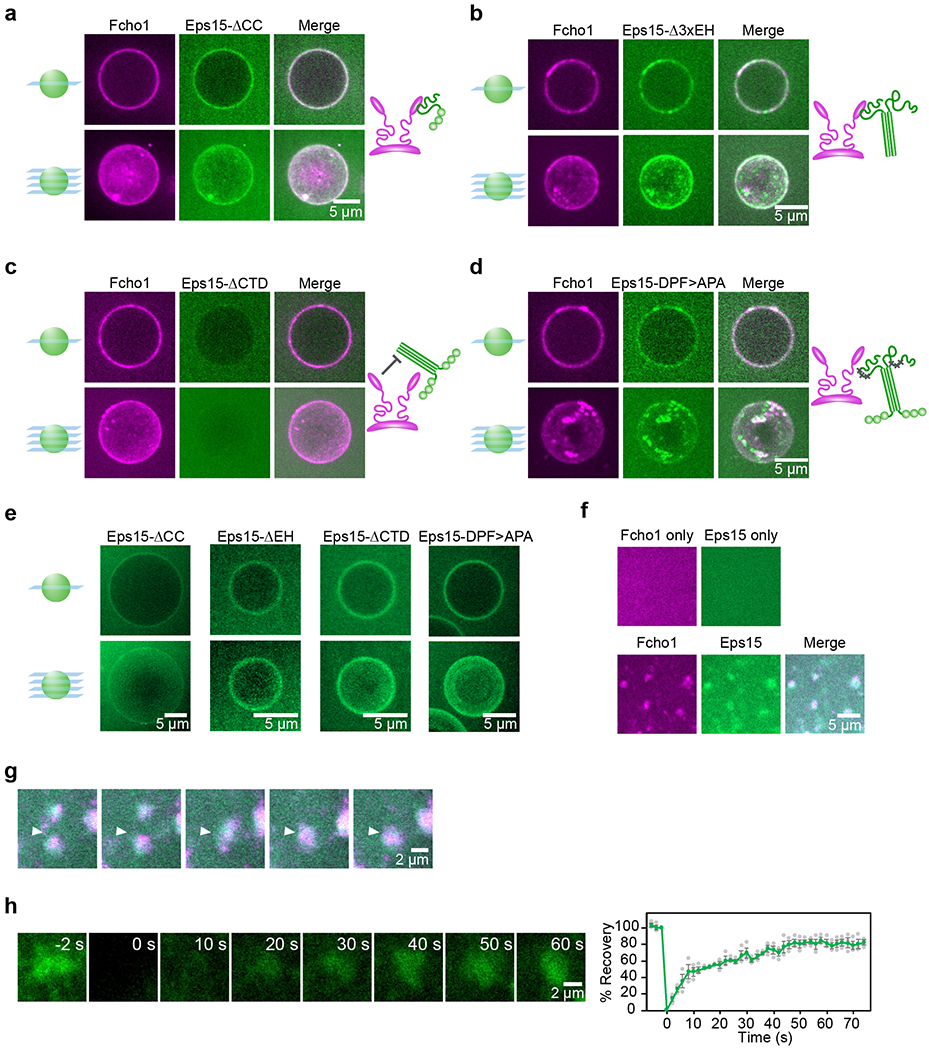
(a-e) 500 nM of the indicated proteins were incubated with GUVs containing 79% DOPC, 15% DOPS, 5% PtdIns(4,5)P2, and 1% DPEG10-biotin. Upper panels display single center z-slices, lower panels display maximum-projected z-stacks. Fcho1 is labeled with Atto594. 6xHis-Eps15 variants are labeled with CF488a. Cartoons depict binding interaction between Fcho1 and Eps15 mutants. (a) GUVs incubated with Fcho1 and Eps15 lacking the coiled-coil domain (Eps15-ΔCC). (b) Fcho1 and Eps15 lacking the EH domains (Eps15-Δ3xEH) occasionally co-assemble into small protein-rich domains. (c) Eps15 lacking the C-terminal disordered domain (Eps15-ΔCTD) is not recruited to GUV surfaces by Fcho1. (d) Eps15 containing mutated Fcho1-binding DPF motifs (amino acids 623-636; Eps15-DPF>APA) co-assembles with Fcho1 into small protein-rich domains. (e) Without Fcho1, Eps15 mutants decorate GUVs homogeneously. GUVs contain 97% DOPC, 2% DOGS-NTA-Ni, 1% DPEG10-biotin. (f-h) Multibilayers containing 73% DOPC, 25% DOPS, and 2% DOGS-NTA-Ni were incubated with 100 nM Atto594-labeled Fcho1, or 100 nM CF488a-labeled Eps15, or both. (f) Fcho1 and Eps15 individually decorate multibilayers homogeneously. When combined, Fcho1 and Eps15 form micron-scale protein-rich regions. (g) Time course of protein-rich Eps15/Fcho1 domains merging on a multibilayer, 6 s intervals. (h) Representative images and plot of fluorescence recovery after bleaching Eps15 (green)/Fcho1 (unlabeled) protein domains on multibilayers. n=3 biologically independent samples. All experiments were conducted at room temperature, 22°C. Scale bars are 5 μm (a-f), or 2 μm (g-h). Data are presented as mean ± SEM. See Source Data Figure 2.
