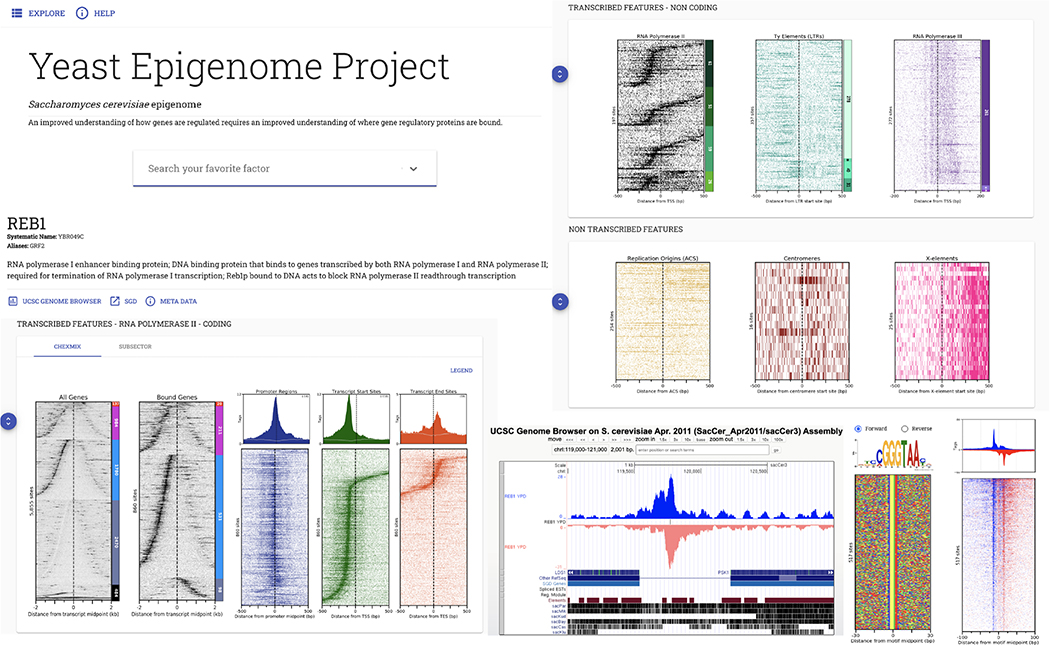
Extended Data Fig. 2 |. Yeastepigenome.org data visualization and discovery.
Shown is an example web browser view at yeastepigenome.org of ChIP-exo occupancy patterns for all targets (e.g., Reb1) around pre-defined genomic features. Rows are sorted by gene or promoter (NFR/NDR) length, or by distance from the indicated reference feature (where x = 0). Promoter classes include (from top to bottom) RP, STM, TFO, UNB, and others. See Supplementary Data 11G,J,C for respective row feature ID, coordinates, and sort order of features that are constant in all target display windows. Lower right (when present) provides strand-separated tag 5’ ends distributed around the protein’s cognate DNA motif, with the motif opposite strand (red) inverted in the composite plot. Corresponding color-coded nucleotide sequences are shown. All images, underlying data values, and datasets can be downloaded through embedded “META DATA” target-specific links at yeastepigenome.org. Each dataset download includes a ReadMe file describing the contents of the download. Warning: Targets with only a single replicate did not pass our significance threshold. See Supplementary Data 11C Internal ID for sort orders that are not provided in the download.