INTRODUCTION:
The prevalence and shedding of fecal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA indicate coronavirus disease 2019 (COVID-19) infection in the gastrointestinal (GI) tract and likely infectivity. We performed a systemic review and meta-analysis to evaluate the prevalence and the duration of shedding of fecal RNA in patients with COVID-19 infection.
METHODS:
PubMed, Embase, Web of Science, and Chinese databases Chinese National Knowledge Infrastructure and Wanfang Data up to June 2020 were searched for studies evaluating fecal SARS-CoV-2 RNA, including anal and rectal samples, in patients with confirmed COVID-19 infection. The pooled prevalence of fecal RNA in patients with detectable respiratory RNA was estimated. The days of shedding and days to loss of fecal and respiratory RNA from presentation were compared.
RESULTS:
Thirty-five studies (N = 1,636) met criteria. The pooled prevalence of fecal RNA in COVID-19 patients was 43% (95% confidence interval [CI] 34%–52%). Higher proportion of patients with GI symptoms (52.4% vs 25.9%, odds ratio = 2.4, 95% CI 1.2–4.7) compared with no GI symptoms, specifically diarrhea (51.6% vs 24.0%, odds ratio = 3.0, 95% CI 1.9–4.8), had detectable fecal RNA. After loss of respiratory RNA, 27% (95% CI 15%–44%) of the patients had persistent shedding of fecal RNA. Days of RNA shedding in the feces were longer than respiratory samples (21.8 vs 14.7 days, mean difference = 7.1 days, 95% CI 1.2–13.0). Furthermore, days to loss of fecal RNA lagged respiratory RNA by a mean of 4.8 days (95% CI 2.2–7.5).
DISCUSSION:
Fecal SARS-CoV-2 RNA is commonly detected in COVID-19 patients with a 3-fold increased risk with diarrhea. Shedding of fecal RNA lasted more than 3 weeks after presentation and a week after last detectable respiratory RNA.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) coronavirus disease 2019 (COVID-19) was first identified in Wuhan, China, in 2019 (1). With rapidly increasing number of patients with SARS-CoV-2 infection worldwide, the World Health Organization (WHO) categorized the spread of COVID-19 to be an international public health emergency and a global pandemic. By September 2020, nearly 27 million confirmed cases and 900,000 deaths worldwide were reported (2). The high infectivity and morbidity of COVID-19 has led to major disruption and even a crisis of healthcare resources in certain epicenters (3). Fever and respiratory symptoms are the most common manifestations of COVID-19, and SARS-CoV-2 is transmitted person to person primarily through respiratory route (4). According to the WHO guidelines, COVID-19 infection is diagnosed by presence of SARS-CoV-2 RNA in respiratory sample by real-time reverse transcription polymerase chain reaction (5,6). However, given the similarity in viral structure with SARS which is in the same family of coronavirus that cause enteric disease, possible pathogenicity of SARS-CoV-2 in the gastrointestinal (GI) tract have been raised. In a meta-analysis of 6,686 patients with COVID-19 infection, the pooled prevalence of GI symptoms was 15% (95% confidence interval [CI] 10%–21%) (7). Furthermore, increasing number of studies demonstrated detection of SARS-CoV-2 RNA in fecal samples (8) and persistent shedding of fecal RNA after recovery from respiratory illness (9). The possibility of transmission of SARS-CoV-2 through fecal-oral route has raised concerns.
Whether GI symptoms in patients with COVID-19 with enteric disease are associated with the presence of fecal RNA is uncertain. Furthermore, the prevalence and duration of viral shedding in the GI tract, even after the loss of virus in the respiratory tract, are unclear. Understanding the natural history of COVID-19 infection in the GI tract is important for optimal public health containment measures. Particularly, concerns for safety of endoscopic services as aerosolizing procedures with increased exposure to fecal material have led to dramatic reductions in procedure volumes (10). We performed a systematic review and meta-analysis to summarize the prevalence and predictors of fecal SARS-CoV-2 RNA. Furthermore, we aimed to compare the duration of viral shedding in fecal and respiratory specimens of patients with COVID-19 infection.
METHODS
Search strategy and selection criteria
In this systematic review and meta-analysis, PubMed, Embase, and Web of Science were searched according to Meta-analysis of Observational Studies in Epidemiology guideline to June 1, 2020. Search strategy terms included: 2019-nCoV-2, coronavirus, COVID-19, SARS-CoV-2, novel coronavirus, or SARS2; and stool, fecal, faecal, rectal, anal, anus, viral RNA, or viral load. Considering the first outbreak in Wuhan, China, 2 major search databases from China (Chinese National Knowledge Infrastructure and Wanfang Data) were searched and reviewed for additional studies in Chinese manuscripts that met criteria.
Studies that met following criteria were eligible for analysis: (i) laboratory confirmed adults and children with respiratory COVID-19 infection and (ii) evaluation of stool collection or anal and rectal swabs for SARS-CoV-2 RNA. We excluded studies evaluating infants, pregnant women, asymptomatic patients, as well as patients with cancer, immune disorders, and other chronic viral infection. All selected literatures were initially screened by title and abstract to exclude irrelevant studies. Full manuscripts of remaining studies, including studies in Chinese, were reviewed to assess for study eligibility. Given the potential inaccuracy, we excluded small studies (N < 10). Two reviewers (Y.W.Z. and M.J.H.) independently assessed for study eligibility, and disagreement was resolved by consensus after discussion with the principal investigators (N.D. and J.K.).
Data extraction and definitions
Information including first author, study date, country of origin, number of patients with fecal or anal/rectal swab SARS-CoV-2 evaluation, number of patients with positive fecal SARS-CoV-2 RNA before and after the loss of respiratory RNA, and duration of positive SARS-CoV-2 RNA in fecal or respiratory samples from the onset of symptoms or hospitalization were extracted. Furthermore, demographic and clinical information including age, sex, disease severity, and symptoms (fever, cough, diarrhea, or other GI symptoms) were collected. GI symptoms were defined by presence of anorexia, nausea, vomiting, abdominal pain/discomfort, GI bleeding, and/or diarrhea. Persistent shedding was defined as the duration from presentation (onset of symptoms or hospitalization) to the last day of positive fecal or respiratory RNA results. Loss of fecal or respiratory RNA was defined as the duration from presentation to the first day of negative fecal or respiratory RNA without a recurrent positive RNA. Patients who lacked documentation of final negative RNA results were excluded for the analysis calculating days to loss of RNA. Severe COVID-19 infection was defined by meeting 1 of the following criteria: (i) respiratory distress with respiratory rate over 30 times per minute, (ii) hypoxia (SpO2 ≤93%) in the resting state, (iii) abnormal blood gas analysis (PaO2/FiO2 ≤300 mm Hg), (iv) severe disease complications including respiratory failure or other organ failure, (v) need of intensive care unit admission, and/or (vi) death (5).
Quality assessment
Quality of enrolled studies were assessed by Agency for Healthcare Research and Quality checklist (http://www.ncbi.nlm.nih.gov/books/NBK35156/). Score of 1 (yes) or 0 (no or unclear) point for each item in the checklist (except for the fifth question with opposite result to other questions) was assigned. Overall score (range of 0–11) was calculated for each study and categorized as high (>7), moderate (4–7), or low (<3) quality.
Data synthesis and statistical analysis
The mean proportions of positive SARS-CoV-2 RNA in feces or anal/rectal swab in patients with confirmed COVID-19 disease and after loss of RNA in respiratory samples were pooled and estimated with 95% CI. The durations of positive SARS-CoV-2 RNA in feces or respiratory samples were estimated by weighted mean difference using the values of the mean and SD. For the studies without mean, we used the value of median, range, and IQR to calculate the mean and SD (11,12). Odds ratios (ORs) were derived to describe the ratio of probability of positive cases with SARS-CoV-2 RNA in feces, occurring in subgroups stratified by age, COVID-19 disease severity, and symptoms presentation (GI symptoms, fever, or cough). Forest plots and funnel plots were used to express the results. Heterogeneity was assessed by I2 statistic. The random-effects model was performed when I2 > 50%, otherwise the fixed-effects model was used. Egger tests were performed to assess for publication bias. Two-sided P value <0.05 was considered statistically significant. All statistical analyses were performed using RStudio version 1.1.463 and Stata SE 15.0 for Mac.
RESULTS
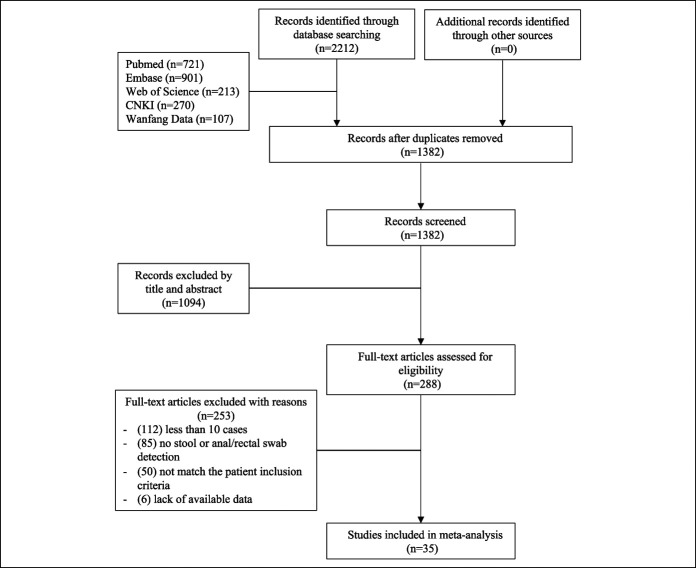
Of 2,212 citations identified, we removed 830 duplicate articles and 1,094 articles after reviewing titles and abstracts. Full-text of 288 relevant articles were reviewed. Finally, 35 studies including 1,636 laboratory confirmed COVID-19 patients who received fecal, anal, and/or rectal swab SARS-CoV-2 RNA examination were included in the analyses (Figure 1). Five studies provided English abstracts with full manuscripts available only in Chinese (13–17).
Figure 1.
Flow diagram of study selection. CNKI, China National Knowledge Infrastructure.
Study characteristics and quality
Of 35 studies, 32 were performed in China (2 in Hong Kong and 1 in Macao), 1 in Italy, 1 in the United States, and 1 in Austria (Table 1). The age of the patients ranged from 8 days to 96 years, and 30%–81% were males. Thirteen studies included patients with both severe and nonsevere disease (15,18–29), 8 included only nonsevere disease (30–37), 3 included only severe disease (16,38,39), and 11 did not specify the disease severity of COVID-19 infection (13,14,17,40–47). The pooled-proportion of patients with severe disease was 35.9% (95% CI 21%–50.9%) in 13 studies evaluating all disease severity types. The median quality score of included studies was 6 (range, 3–8) indicating moderate quality.
Table 1.
Characteristics of included studies
| Study | Publish or accepted date | Study period | Region | N | Age | Male | Disease severity | Quality score |
| Chen et al. (18) | February 15, 2020 | NA | China | 28 | NA | NA | 12 severe; 16 nonsevere | 4 |
| Chen et al. (19) | March 31, 2020 | January 20, 2020–February 9, 2020 | China | 42 | Median 51 (IQR 43–62) | 15 (35.7%) | 11 severe; 31 nonsevere | 5 |
| Cheung et al. (40) | March 26, 2020 | February 2, 2020 – February 29, 2020 | Hong Kong | 59 | Median 58.5 (IQR 44–68, range 22–96) | 27 (45.8%) | NA | 5 |
| De Ioris et al. (41) | May 23, 2020 | March 16, 2020 – April 8, 2020 | Italy | 22 | Median 84 mo (8 d–210 mo) | 15 (68.2%) | NA | 6 |
| Effenberger et al. (42) | April 13, 2020 | NA | Austria | 40 | Without diarrhea: 58.4 ± 17.1; resolved diarrhea: 66.3 ± 13.1; with diarrhea: 78.3 ± 13.8 | 24 (60%) | NA | 5 |
| Han et al. (30) | March 31, 2020 | February 13, 2020 – February 29, 2020 | China | 22 | 43.3 ± 14.2 | 9 (40.9%) | Nonsevere type | 6 |
| Huang et al. (38) | April 15, 2020 | January 26, 2020–February 25, 2020 | China | 16 | Median 60 (range 26–79) | 13 (81.3%) | Severe type to ICU | 6 |
| Kujawski et al. (20) | April 23, 2020 | January 20, 2020–February 5, 2020 | US | 12 | Median 53 (range 21–68) | 8 (66.7%) | All types | 4 |
| Li et al. (13) | February 17, 2020 | February 11, 2020–February 13, 2020 | China | 15 | NA | NA | NA | 5 |
| Li et al. (31) | April 20, 2020 | January 26, 2020–February 6, 2020 | China | 13 | 52.8 ± 20.2 (range 1–72) | 6 (46.2%) | Nonsevere type | 6 |
| Lin et al. (21) | March 24, 2020 | January 17, 2020–February 15, 2020 | China | 65 | 45.3 ± 18.3 | NA | All types | 6 |
| Ling et al. (43) | May 5, 2020 | January 20, 2020–February 10, 2020 | China | 66 | Median 44 (IQR 34–62, range 16–78) | 38 (57.6%) | NA | 7 |
| Liu et al. (44) | April 14, 2020 | March 15, 2020 | China | 69 | NA | NA | NA | 7 |
| Lo et al. (22) | March 15, 2020 | January 21, 2020–February 16, 2020 | Macau | 10 | Median 54 (IQR 27–64) | 3 (30%) | 6 severe; 4 nonsevere | 7 |
| Ma et al. (32) | March 19, 2020 | NA | China | 27 | NA | NA | Nonsevere type | 3 |
| Tan et al. (33) | April 3, 2020 | January 27, 2020–March 10, 2020 | China | 10 | Mean 7 (range 1–12) | 3 (30%) | Nonsevere type | 7 |
| To et al. (23) | March 23, 2020 | January 22, 2020–February 12, 2020 | Hong Kong | 15 | Median 62 (range 37–75) | NA | 8 severe; 7 nonsevere | 7 |
| Wang et al. (14) | April 21, 2020 | January 24, 2020–February 17, 2020 | China | 50 | 42.6 ± 17.5 | 24 (48%) | NA | 7 |
| Wang et al. (24) | March 11, 2020 | January 1, 2020–February 17, 2020 | China | 16 | NA | NA | All types | 6 |
| Wei et al. (34) | May 25, 2020 | January 19, 2020–February 7, 2020 | China | 84 | Median 37 (range 24–74) | 28 (33%) | Nonsevere type | 7 |
| Wu et al. (15) | February 27, 2020 | NA | China | 36 | 49 (range 17–86) | 22 (61.1%) | 12 severe; 24 nonsevere | 4 |
| Wu et al. (16) | March 31, 2020 | January 31, 2020–February 19, 2020 | China | 19 | Mean 67 (range 51–86) | 10 (52.6%) | 17 severe; 2 critical severe | 6 |
| Wu et al. (25) | March 19, 2020 | January 16, 2020–March 15, 2020 | China | 74 | Stool RNA+ patients: 41.3 ± 3.1; stool RNA− patients: 46.2 ± 2.6 | 39 (52.7%) | 18 severe; 56 nonsevere | 7 |
| Xiao et al. (39) | February 27, 2020 | February 1, 2020–February 14, 2020 | China | 73 | Mean 43 (range 0.8–78 yr) | 41 (56.2%) | 4 ICU | 6 |
| Xu et al. (35) | March 7, 2020 | January 23, 2020–February 18, 2020 | China | 51 | Imported cases: median 35 (IQR 29–51) Secondary cases: median 37 (IQR 24–48) Tertiary cases: median 53 (IQR 35–65) |
25 (49.0%) | Nonsevere type | 7 |
| Xu et al. (36) | April 2020 | By February 20, 2020 | China | 10 | Range: 2 mo–15 yr | 6 (60%) | Nonsevere type | 6 |
| Yuan et al. (45) | May 18, 2020 | January 1, 2020–March 18, 2020 | China | 61 | NA | NA | NA | 8 |
| Zhang et al. (37) | May 15, 2020 | January 17, 2020–January 28, 2020 | China | 22 | NHM group: 43.4 ± 15.9; control group: 40.7 ± 13.3 | 8 (36.4%) | Nonsevere type | 8 |
| Zhang et al. (46) | March 1, 2020 | January 27, 2020–February 10, 2020 | China | 14 | Median 41 (range 18–87) | 7 (50%) | NA | 6 |
| Zhang et al. (26) | May 2, 2020 | January 25, 2020–March 18, 2020 | China | 15 | Median 37 (range 10–73) | 8 (53.3%) | 4 severe; 8 nonsevere; 3 asymptomatic | 7 |
| Zhao et al. (27) | May 12, 2020 | NA | China | 401 | Median 47 (IQR 33–60) | 190 (47.4%) | 85 severe; 316 nonsevere | 7 |
| Zheng et al. (17) | April 24, 2020 | January 30, 2020–February 23, 2020 | China | 51 | Male: 42.6 ± 16.11; female: 38.3 ± 19.03 | 25 (49.0%) | NA | 6 |
| Zheng et al. (47) | April 20, 2020 | January 25, 2020–February 26, 2020 | China | 20 | Range: 23–57 yr | 14 (70%) | NA | 6 |
| Zheng et al. (28) | April 6, 2020 | January 19, 2020–February 15, 2020 | China | 93 | Median 55 (IQR 44.3–64.8) | NA | 71 severe; 22 nonsevere | 6 |
| Zuo et al. (29) | May 14, 2020 | February 5, 2020–March 17, 2020 | China | 15 | Median 55 (IQR 44–67.5) | 7 (47%) | All types | 8 |
IQR, interquartile range; NA, no description.
Prevalence of fecal SARS-CoV-2 RNA
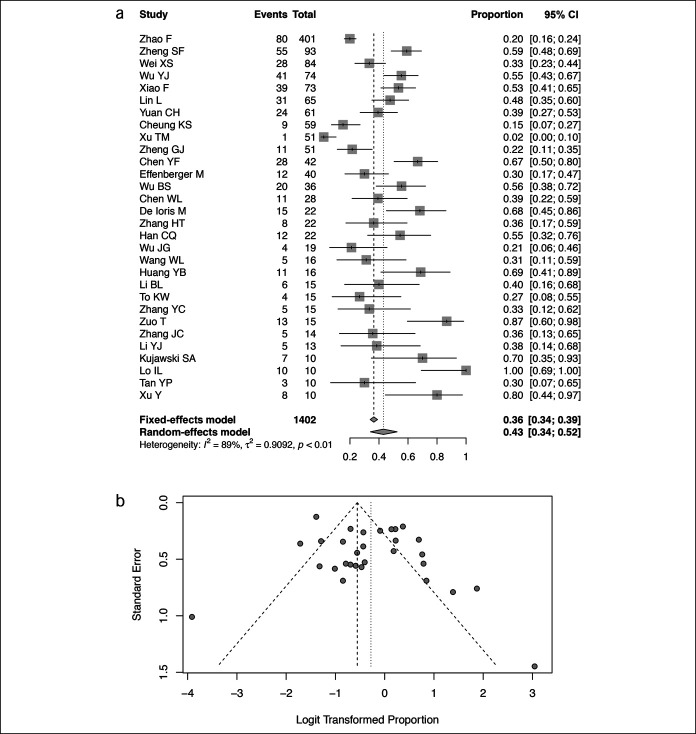
Thirty studies reported the prevalence of fecal SARS-CoV-2 RNA in patients with COVID-19 infection confirmed by respiratory samples (13,15–31,33–35,37–42,45,46). The pooled prevalence was 43% (95% CI 34%–52%, I2 = 89%) (Figure 2a). The highest prevalence was 100% in a study from Macau (22), and the lowest was 2% in a study evaluating patients with severe disease (35). The funnel plot suggested absence of publication bias based on the Egger test (P = 0.10, Figure 2b). Subgroup analysis showed no differences in the prevalence of fecal RNA comparing adults and children (39% vs 54%, P = 0.22) or patients with nonsevere and severe disease (35% vs 49%, P = 0.21) (see Supplemental Figure, Supplementary Digital Content 1, http://links.lww.com/CTG/A570).
Figure 2.
Pooled prevalence of detectable SARS-CoV-2 RNA in fecal samples of patients with confirmed COVID-19 infection. (a) Forest plots of included studies. (b) Funnel plots of included studies. CI, confidence interval; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Prevalence of fecal SARS-CoV-2 RNA by symptoms
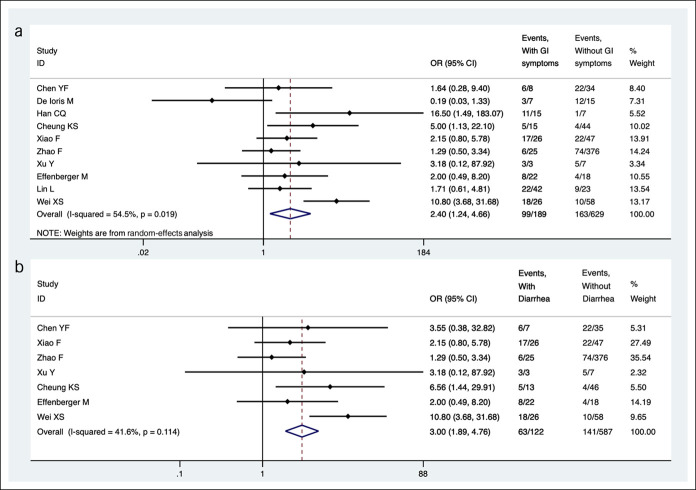
Sixteen studies compared the prevalence of fecal SARS-CoV-2 RNA in patients with different clinical symptoms (15,18,19,21,23,25–28,30,34,36,39–42). Pooled results from 10 studies indicated that higher proportion of patients had detectable fecal SARS-CoV-2 RNA among those with GI symptoms (52.4% [99/189] vs 25.9% [163/629]; OR = 2.4, 95% CI 1.2–4.7, I2 = 54.5%) compared with no GI symptoms (Figure 3a). Patients with diarrhea were more likely to test positive for fecal SARS-CoV-2 RNA (51.6% [63/122] vs 24.0% [141/587], OR = 3.0, 95% CI 1.9–4.8, I2 = 41.6%) compared with those without diarrhea (Figure 3b). In subgroup analysis, no differences in odds of detectable fecal RNA were observed among patients with severe or nonsevere disease (OR = 1.2, 95% CI 0.8–1.7) (15,18,19,23,25–28,39), with or without a fever (OR = 1.0, 95% CI 0.7–1.6) (19,25,27,30,36), and with or without cough (OR = 1.1, 95% CI 0.7–1.7) (19,25,27,36).
Figure 3.
Comparison of pooled prevalence of SARS-CoV-2 RNA in fecal samples of patients with COVID-19 infection with or without GI symptoms. (a) Comparison among patients with or without GI symptoms. (b) Comparison among patients with or without diarrhea. CI, confidence interval; COVID-19, coronavirus disease 2019; GI, gastrointestinal; OR, odds ratio; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Persistent shedding of fecal SARS-CoV-2 RNA
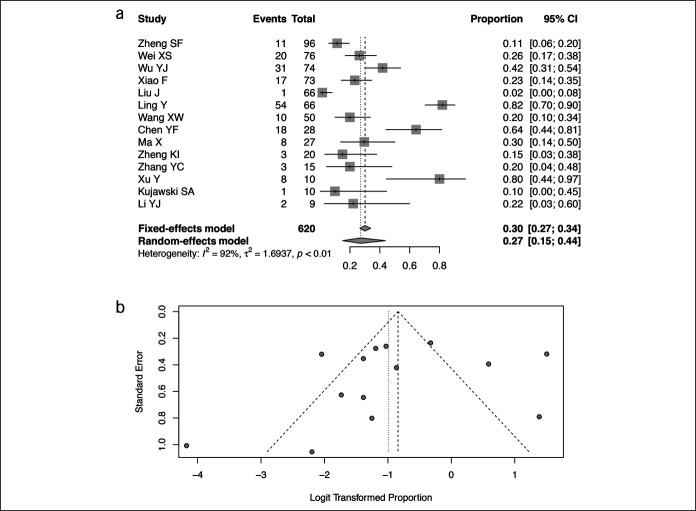
Fourteen studies including 620 patients had available data on the prevalence of SARS-CoV-2 RNA in feces after virus converted negative in respiratory samples (Figure 4a) (14,19,20,25,26,28,31,32,34,36,39,43,44,47). The pooled prevalence was 27% (95% CI 15%–44%, I2 = 92%). No publication bias was observed (P = 0.50, Figure 4b).
Figure 4.
Pooled prevalence of SARS-CoV-2 RNA in fecal samples of patients with COVID-19 patients after loss of RNA in respiratory samples. (a) Forest plots of included studies. (b) Funnel plots of included studies. CI, confidence interval; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
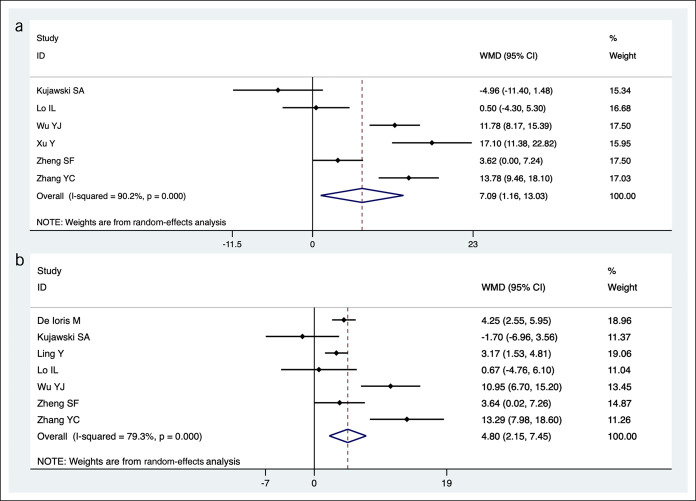
Duration of persistent shedding of SARS-CoV-2 RNA in feces or respiratory samples was assessed in 8 studies (Figure 5) (20,22,25,26,28,36,41,43). The pooled mean durations of persistent shedding from the onset of symptoms to the final documented detectable viral RNA were 21.8 days (95% CI 16.4–27.1) in feces and 14.7 days (95% CI 9.9–19.5) in respiratory samples with a mean difference of 7.1 days (95% CI 1.2–13.0, Figure 5a). Furthermore, days to loss of RNA was longer in fecal (19.9 days, 95% CI 15.6–24.1) compared with respiratory samples (15.0 days, 95% CI 10.9–19.2) with a mean difference of 4.8 days (95% CI 2.2–7.5, Figure 5b).
Figure 5.
Comparison of pooled durations for persistent shedding and loss of SARS-CoV-2 RNA in fecal and respiratory samples of patients with COVID-19 infection. (a) Persistent shedding of SARS-CoV-2 RNA in fecal/anal/rectal and respiratory samples of COVID-19 patients. (b) Loss of SARS-CoV-2 RNA in fecal/anal/rectal and respiratory samples of COVID-19 patients. Results of respiratory samples were used as control during comparison. CI, confidence interval; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WMD, weighted mean difference.
DISCUSSION
We performed a systematic review and meta-analysis of 1,636 patients to evaluate the prevalence and the duration of shedding of fecal RNA in patients with COVID-19 infection. Overall, 43% (95% CI 34%–52%) of patients with COVID-19 infection tested positive for fecal RNA with a higher proportion observed in patients with GI symptoms, especially diarrhea, compared with no GI symptoms. After loss of RNA in the respiratory tract, 27% (95% CI 15%–44%) had persistent detectable RNA in the feces. Furthermore, SARS-CoV-2 RNA was detectable in the feces for a mean of 21.8 days (95% CI 1.2–13.0) from presentation with a mean difference of 7.1 days (95% CI 1.2–13.0) greater than the last detectable virus in the respiratory tract. Similarly, loss of fecal RNA lagged the loss of RNA in the respiratory tract by mean of 4.8 days (95% CI 2.2–7.5).
Emerging studies revealed that SARS-CoV-2 possesses high degree of homology with SARS-CoV in the family of B beta-coronavirus and may share similar pathogenicity (48). Previous studies in SARS showed that >60% of infected patients had detectable positive fecal viral RNA which could last >4 weeks even after discharge from hospitalization (49,50). Similarly, we found that COVID-19 patients also showed a high prevalence of fecal viral RNA and prolonged viral shedding in feces. Some studies reported protracted shedding of fecal RNA of 2 months even after complete recovery of respiratory disease and loss of respiratory RNA (28). Our meta-analysis showed that 27% of patients who cleared the virus from the respiratory tract with 2 consecutive negative tests had detectable fecal RNA. A recent systemic review that included both suspected and confirmed respiratory COVID-19 infection also showed a high prevalence (64%) of detectable fecal RNA and a protracted fecal viral shedding after loss of RNA in respiratory samples (mean difference of 12.5 days) (51). A higher proportion of patients with persistent shedding of fecal RNA after loss of respiratory RNA in the previous study may be related to indiscriminately including patients with positive fecal SARS-CoV-2 RNA regardless of meeting WHO criteria of COVID-19 infection that require a positive respiratory RNA. In clinical practice, fecal RNA alone is seldomly evaluated in patients with suspected COVID-19 infection. To increase generalizability, the prevalence of positive fecal RNA and persistent shedding of fecal RNA were calculated only among patients with confirmed COVID-19 infection based on WHO criteria in our study.
Angiotensin-converting enzyme II (ACE2) and transmembrane serine protease 2 (TMPRSS2) are 2 main cell entry receptors of SARS-CoV for host invasion. Not only in lung alveolar type 2 cells, ACE2 and TMPRSS2 are also highly expressed in the GI tract including stomach, ileum, and colon based on single-cell transcriptomic analysis (52). As a prerequisite for SARS-CoV-2 infection, the abundant distribution of ACE2 and TMPRSS2 in GI epithelium provides opportunity for viral host entry. Given the high prevalence of fecal RNA in patients with COVID-19 patients, the GI tract is a likely target organ of SARS-CoV-2. Parallel with the proposed mechanism, the digestive symptoms are common in COVID-19 patients. Previous meta-analyses estimated that 15% of patients with COVID-19 infection exhibit GI symptoms (7,40). Our analysis also showed that presence of GI symptom increased the odds of detectable fecal SARS-CoV-2 RNA (52% vs 26%, OR = 2.4, 95% CI 1.2–4.7), supporting a strong association between presence of GI symptoms and fecal SARS-CoV-2 in patients with COVID-19 infection. In addition to fecal samples, SARS-CoV-2 RNA and proteins have been isolated from GI tract (21,39). Furthermore, infectious SARS-CoV-2 has been successfully isolated from fecal samples (21). Currently, pathogenicity and mechanism of SARS-CoV-2 on the GI tract are largely unknown. However, preliminary studies demonstrated that patients with COVID-19 exhibit gut dysbiosis characterized by enrichment of opportunistic pathogens, loss of short-chain fatty acid–producing bacteria, and increased functional capacity for nucleotide, amino acid, and carbohydrate metabolism (53). The interaction between SARS-CoV-2 and gut commensal bacteria leading to microbial shifts may account for the development of GI symptoms in patients with COVID-19 infection. Further studies are needed to elucidate the pathogenicity of SARS-CoV-2 in the GI tract.
Although early evidence suggested that fecal-oral transmission SARS-CoV-2 may be possible, a case of direct feces-oral transmission is yet to be reported (54). Several studies showed that SARS-CoV-2 can be detected in feces-related contaminants, such as wastewater, toilet plumes, and hospital sewage system (55–57). Whether the isolated virus from feces-related contaminants have viability or infectivity is unclear. Although culturing the virus from feces is possible, attempts to culture the virus from feces-related contaminants (i.e., sewage) have not been successful (56). Future studies examining the viability and infectivity of SARS-CoV-2 virus from feces-related contaminants will be important to clarify the significance of fecal shedding of SARS-CoV-2. In addition, emerging studies have demonstrated the potential application of waste water-based epidemiology to track the spread of COVID-19 infection at the population level (58). For example, longitudinal quantification of SARS-CoV-2 RNA by reverse-transcription quantitative polymerase chain reaction of waste water treatment plant samples in France correlated with both the rise and decline in the number of new cases (59). Another study from Spain showed that longitudinal sampling of waste water provided early evidence of circulating COVID-19 infection 12–16 days before the first clinical case within the treatment plant catchment area (60). Finally, a US study demonstrated that the rise in SARS-CoV-2 RNA concentrations in sewage sludge preceded the rise in the number and proportion of positive test results by 0–2 days and the rise in the number of local hospital admission by 1–4 days (61). The ability to accurately assess the burden of COVID-19 infection at the population level will be particularly important, not only for targeting quarantine measures but also appropriating mass vaccination efforts for optimal disease control currently underway (62).
Our findings have clinical implications on public health policies. Given our results showing high prevalence of fecal RNA and prolonged shedding of virus 3 weeks from presentation, heightened level of caution is warranted for patients with GI COVID-19 infection. Currently, the WHO guidelines recommend quarantine of 10 days and the Centers for Disease Control recommend 14 days after onset of primarily respiratory symptoms without laboratory testing (63,64). Fecal RNA testing is not routinely performed in clinical practice, and the presence of GI symptoms is not incorporated in policies for quarantine measures. Given that the final loss of fecal RNA was not documented in many of the studies, persistent shedding of fecal RNA likely exceeded 3 weeks (36). Therefore, a longer duration of quarantine (i.e., 3–4 weeks) should be considered among those with or at risk of GI COVID-19 infection (especially diarrhea) until the risk of fecal-oral transmission is fully clarified. In addition, our findings also may impact endoscopy practices. Although dramatic reduction in endoscopy volumes was commonly observed at the start of the pandemic (1), many centers have cautiously resumed performing elective procedures (65). Society guidelines recommend COVID-19 before testing in respiratory samples 48–72 hours before elective endoscopy where the prevalence of asymptomatic SARS-CoV-2 infection is intermediate (0.5%–2%) (66). Increased number of positive results in asymptomatic patients from routine pretesting for COVID-19 infection is expected. Furthermore, when to safely perform endoscopic procedures in hospitalized patients with COVID-19 infection is uncertain. Currently, there are no guidelines to decide when or how to perform semi-urgent endoscopy (e.g., management of inflammatory bowel disease and cancer treatment) in patients with COVID-19 infection. However, based on our results, delaying endoscopic procedures for >3–4 weeks may be warranted unless medical urgency necessitates an earlier procedure.
Strengths of our study include analysis of a large number of patients (N = 1,636) with GI COVID-19 infection based on fecal SARS-CoV-2 RNA, quadrupling the sample size of the previous meta-analysis (67). Our study is also first to estimate the prevalence of detectable fecal SARS-CoV-2 RNA after the loss of RNA in respiratory samples in patients with laboratory-confirmed COVID-19 infection. Finally, literature search and full review of Chinese manuscripts without limitation were possible by the investigators because several studies were only available in Chinese language.
The current study has limitations. First, the quality of the observational studies included in the meta-analysis was not high. Substantial heterogeneity was present among the studies likely reflecting differences in testing methods for SARS-CoV-2 RNA, definitions of positive samples, and patient population. Furthermore, findings of the meta-analysis in predominantly Chinese population may limit the generalizability in other setting. Finally, asymptomatic patients with COVID-19 infection were excluded given the paucity of studies and will be invaluable in future studies.
In conclusion, fecal SARS-CoV-2 RNA was commonly detected in patients with COVID-19 patients, exceeding half among those with GI symptoms. More than a quarter of patients had prolonged shedding of fecal RNA after clearance of RNA in the respiratory tract, and fecal RNA was detected for 3 weeks after presentation, exceeding the duration of shedding in the respiratory tract by a week. Policies incorporating GI symptoms or evaluation of fecal RNA to reduce the risk of exposure of SARS-CoV-2 RNA to healthcare worker and patients should be considered until infectivity of fecal-oral transmission is fully elucidated.
CONFLICTS OF INTEREST
Guarantor of the article: Ning Dai, MD, PhD, and John J. Kim, MD, MS.
Specific author contributions: Y.W.Z.: collected and interpreted data, drafted the manuscript, and approved the final version of the manuscript. M.S.C.: and M.J.H.: collected data and approved the final version of the manuscript. L.J.D.: drafted the manuscript and approved the final version of the manuscript. W.L.H., J.J.K., and N.D.: planned and conducted the study, interpreted data, and approved the final version of the manuscript.
Financial support: This work was supported by the Medical Health Project of Zhejiang Province (2020RC064).
Potential competing interests: None to report.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A570.
Contributor Information
Yawen Zhang, Email: 11718175@zju.edu.cn.
Mengsha Cen, Email: 21818348@zju.edu.cn.
Mengjia Hu, Email: hu_mengjia@126.com.
Lijun Du, Email: dlj@zju.edu.cn.
Weiling Hu, Email: huweiling@zju.edu.cn.
Ning Dai, Email: ndaicn@zju.edu.cn.
References
- 1.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus Disease (COVID-19): Weekly Epidemiological Update. World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- 3.Khan G, Sheek-Hussein M, Al Suwaidi AR, et al. Novel coronavirus pandemic: A global health threat. Turk J Emerg Med 2020;20:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan WJ, Ni ZY, Hu Y, et al. ; China Medical Treatment Expert Group For Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020;382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (2019-nCoV) Infection Is Suspected: Interim Guidance, 28 January 2020. World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- 6.Laboratory Testing for Coronavirus Disease (COVID-19) in Suspected Human Cases. Interim Guidance 19 March 2020. World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- 7.Mao R, Qiu Y, He JS, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2020;5:667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S, Parker J, Smits S, et al. Persistent viral shedding of SARS-CoV-2 in faeces—A rapid review. Colorectal Dis 2020;22:611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Zhou Y, Jiang N, et al. Persistence of intestinal SARS-CoV-2 infection in patients with COVID-19 leads to re-admission after pneumonia resolved. Int J Infect Dis 2020;95:433–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutter MD, Brookes M, Lee TJ, et al. Impact of the COVID-19 pandemic on UK endoscopic activity and cancer detection: A National Endoscopy Database Analysis. Gut 2020;70:537–43. [DOI] [PubMed] [Google Scholar]
- 11.Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol 2005;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan X, Wang W, Liu J, et al. Estimating the sample mean and standard deviation from the sample size, median, range and:or interquartile range. BMC Med Res Methodol 2014;14:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li BL, Li Q, Wu G, et al. Comparison of 2019-nCoV test results of sputum and fecal specimens of 15 patients with COVID-19 after treatment. Chin J Infect Control 2020;19:239–44. [Google Scholar]
- 14.Wang XW, Zhu YL, Li TT, et al. Evaluation of SARS-COV-2 nucleic acid in convalescent anal swabs of patients with novel coronavirus infection. Tianjin Med J 2020;48:498–501. [Google Scholar]
- 15.Wu BS, Yu TT, Huang ZM, et al. Nucleic acid detection of fecal samples from confirmed cases of COVID-19. Chin J Zoonoses 2020;36:359–361. [Google Scholar]
- 16.Wu JG, Luo JF, Liu JS, et al. Detection of 2019-nCoV by real-time RT-PCR using multiple biological samples in severe/critically ill patients. Acad J Chin PLA Med Sch 2020;41:205–207. [Google Scholar]
- 17.Zheng GJ, Zhang HY, Chen CH, et al. A comparative analysis of nucleic acid test of throat swab and anal swab specimen types from 51 patients with novel coronavirus pneumonia in Changzhou. Chin J Nosocomiol 2020;30:961–3. [Google Scholar]
- 18.Chen W, Lan Y, Yuan X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect 2020;9:469–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Chen L, Deng Q, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol 2020;92:833–40. [DOI] [PubMed] [Google Scholar]
- 20.Kujawski SA, Wong KK, Collins JP, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 2020;26:861–8. [DOI] [PubMed] [Google Scholar]
- 21.Lin L, Jiang X, Zhang Z, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020;69:997–1001. [DOI] [PubMed] [Google Scholar]
- 22.Lo IL, Lio CF, Cheong HH, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci 2020;16:1698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect Dis 2020;20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 2020;5:434–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YC, Han S, Wang XN, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect 2020;9:833–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao F, Yang Y, Wang Z, et al. The time sequences of respiratory and rectal viral shedding in patients with coronavirus disease 2019. Gastroenterology 2020;159:1158–60.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng S, Fan J, Yu F, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020;369:m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020;159:944–55.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han C, Duan C, Zhang S, et al. Digestive symptoms in COVID-19 patients with mild disease severity: Clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol 2020;115:916–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Hu Y, Yu Y, et al. Positive result of Sars-Cov-2 in faeces and sputum from discharged patient with COVID-19 in Yiwu, China. J Med Virol 2020;92:1938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X, Su L, Zhang Y, et al. Do children need a longer time to shed SARS-CoV-2 in stool than adults? J Microbiol Immunol Infect 2020;53:373–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan YP, Tan BY, Pan J, et al. Epidemiologic and clinical characteristics of 10 children with coronavirus disease 2019 in Changsha, China. J Clin Virol 2020;127:104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei XS, Wang X, Niu YR, et al. Diarrhea is associated with prolonged symptoms and viral carriage in corona virus disease 2019. Clin Gastroenterol Hepatol 2020;18:1753–9.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu T, Chen C, Zhu Z, et al. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int J Infect Dis 2020;94:68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Li X, Zhu B, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 2020;26:502–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang HT, Huang MX, Liu X, et al. Evaluation of the adjuvant efficacy of natural herbal medicine on COVID-19: A retrospective matched case-control study. Am J Chin Med 2020;48:779–92. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Chen S, Yang Z, et al. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am J Respir Crit Care Med 2020;201:1435–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020;158:1831–3.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung KS, Hung IF, Chan PP, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology 2020;159:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Ioris MA, Scarselli A, Ciofi Degli Atti ML, et al. Dynamic viral SARS-CoV-2 RNA shedding in in children: Preliminary data and clinical consideration of Italian regional center. J Pediatric Infect Dis Soc 2020;9:366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Effenberger M, Grabherr F, Mayr L, et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut 2020;69:1543–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling Y, Xu SB, Lin YX, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 2020;133:1039–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Xiao Y, Shen Y, et al. Detection of SARS-CoV-2 by RT-PCR in anal from patients who have recovered from coronavirus disease 2019. J Med Virol 2020;92:1769–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan C, Zhu H, Yang Y, et al. Viral loads in throat and anal swabs in children infected with SARS-CoV-2. Emerg Microbes Infect 2020;9:1233–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol 2020;92:680–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng KI, Wang XB, Jin XH, et al. A case series of recurrent viral RNA positivity in recovered COVID-19 Chinese patients. J Gen Intern Med 2020;35:2205–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung IF, Lau SK, Woo PC, et al. Viral loads in clinical specimens and SARS manifestations. Hong Kong Med J 2020;15:20–2. [PubMed] [Google Scholar]
- 50.Xu D, Zhang Z, Jin L, et al. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur J Clin Microbiol Infect Dis 2005;24:165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Doorn AS, Meijer B, Frampton CMA, et al. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Aliment Pharmacol Ther 2020;52:1276–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Kang Z, Gong H, et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020;69:1010–8. [Google Scholar]
- 53.Zuo T, Liu Q, Zhang F, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2020;70:276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel KP, Vunnam SR, Patel PA, et al. Transmission of SARS-CoV-2: An update of current literature. Eur J Clin Microbiol Infect Dis 2020;39:2005–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lodder W, De Roda Husman AM. SARS-CoV-2 in wastewater: Potential health risk, but also data source. Lancet Gastroenterol Hepatol 2020;5:533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Feng H, Zhang S, et al. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int J Infect Dis 2020;94:103–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li YY, Wang JX, Chen X. Can a toilet promote virus transmission? From a fluid dynamics perspective. Phys Fluids (1994) 2020;32:065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rooney CM, Moura IB, Wilcox MH. Tracking COVID-19 via sewage. Curr Opin Gastroenterol 2021;37:4–8. [DOI] [PubMed] [Google Scholar]
- 59.Wurtzer S, Marechal V, Mouchel JM, et al. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill 2020;25:2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Randazzo W, Truchado P, Cuevas-Ferrando E, et al. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res 2020;181:115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peccia J, Zulli A, Brackney DE, et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol 2020;38:1164–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith T, Cassell G, Bhatnagar A. Wastewater surveillance can have a second act in COVID-19 vaccine distribution. JAMA Health Forum 2021;2:e201616. [DOI] [PubMed] [Google Scholar]
- 63.Home Care for Patients With Suspected or Confirmed COVID-19 and Management of Their Contacts. Inteirm Guidance 12 August 2020. World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- 64.Coronavirus Disease 2019 (COVID-19): When to Quarantine. Centers for Disease Control and Prevention: Atlanta, GA, 2020. [Google Scholar]
- 65.Miura F, Kitajima M, Omori R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: Re-analysis of patient data using a shedding dynamics model. Sci Total Environ 2021;769:144549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sultan S, Siddique SM, Altayar O, et al. ; American Gastroenterological Association. AGA institute rapid review and recommendations on the role of pre-procedure SARS-CoV2 testing and endoscopy. Gastroenterology 2020;159:1935–48.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong MC, Huang J, Lai C, et al. Detection of SARS-CoV-2 RNA in fecal specimens of patients with confirmed COVID-19: A meta-analysis. J Infect 2020;81:e31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.