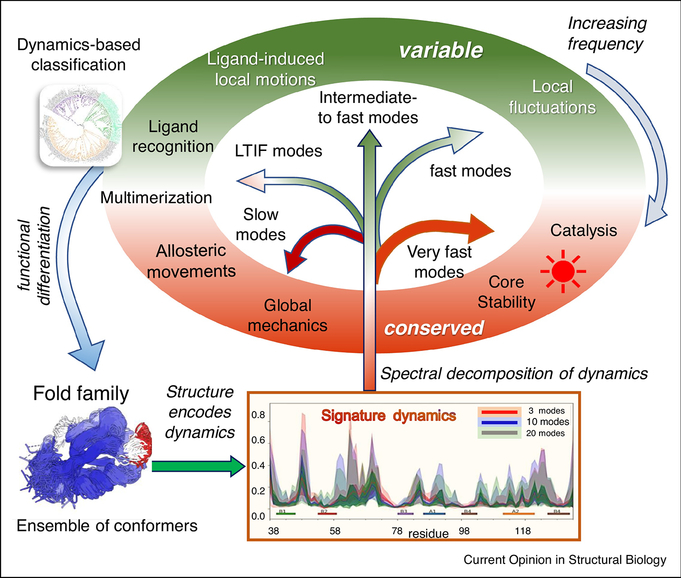
Figure 1: Significance of protein dynamics in enabling functional adaptation and specialization within protein families.
Various events, such as gene duplication and random mutagenesis, give rise to ensembles of homologous protein/domain structures, also called fold families (structurally aligned at the bottom left for PDZ domains). Each family has its unique structure-encoded dynamics under equilibrium conditions, represented by a spectrum of normal modes of motion, and a family-specific signature dynamics (middle bottom for PDZ domains). The proteins whose structures and dynamics are well adapted to do biological functions (in the elliptical shell) are evolutionarily retained/selected and vice versa. Hence, evolution leads to an iterative refinement of protein structure and dynamics to enable fold adaptation and functional specialization (curled blue arrow on the left). An ensemble normal mode analysis allows us to identify and compare the different frequency modes encoded by each structure in the ensemble. Notably, motions at two ends of the spectrum, shortly called global/slowest and very fast modes, are conserved (highlighted in red/brick in the elliptical shell) for diametrically opposed reasons: the former underlies the global mechanics/rearrangements required for adaptation to intermolecular interactions, including ligand binding and allosteric responses; the latter reflects key interactions that cannot tolerate sequence/structure perturbations, such as the highly stable packing at the core of the fold, or specific interactions at chemically active (e.g. catalytic) sites that are precisely retained as the fundamental characteristic feature of the family. LTIF modes usually define subfamilies, that tolerate variations within subfamilies, and drive fold adaptation and functional differentiation. They are conserved within, but vary between, subfamilies. Fast/intermediate modes show maximal variance between members within and across subfamilies (green portions of the ellipse).