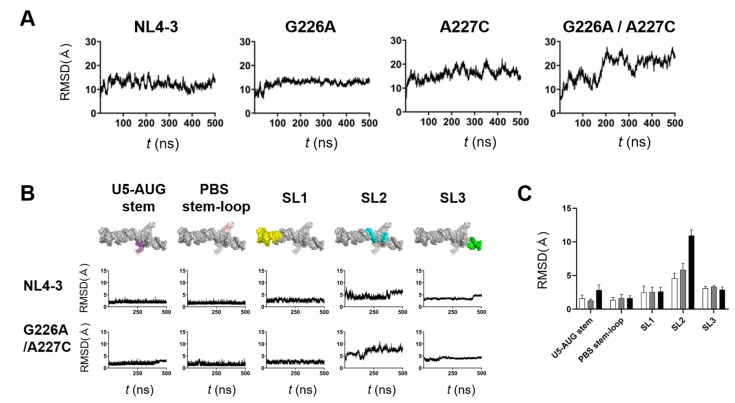
Figure 5.
Effects of the dinucleotide substitution on the structural dynamics of psi. ΨCES models without or with indicated substitution were subjected to MD simulations using the Amber 16 program package [19]. Structural changes during 0–500 ns of MD simulations were monitored with the root mean square deviations (RMSDs) between the initial model structure and the structures at given time points of MD simulation using the cpptraj module in AmberTools 16 as described previously [20,21,22,23,24]. (A) and (B) Changes in RMSDs during MD simulations. ΨCES RNAs without or with indicated substitution (A). Structural units of ΨCES RNAs without or the 226/227 dinucleotide substitution (B). (C) Means of RMSDs of indicated structural units of ΨCES RNA. Structural units of ΨCES were extracted from ΨCESs at 460, 470, 480, 490, and 500ns after MD simulations, superposed, and used for calculating RMSDs between NL4-3 and mutants. White, gray, black bars indicate RMSDs between NL4-3 ΨCES and ΨCESs with a G226A substitution, an A227C substitution, and a G226A/A227C dinucleotide substitution, respectively. Mean values and standard deviation with the five time points are shown.