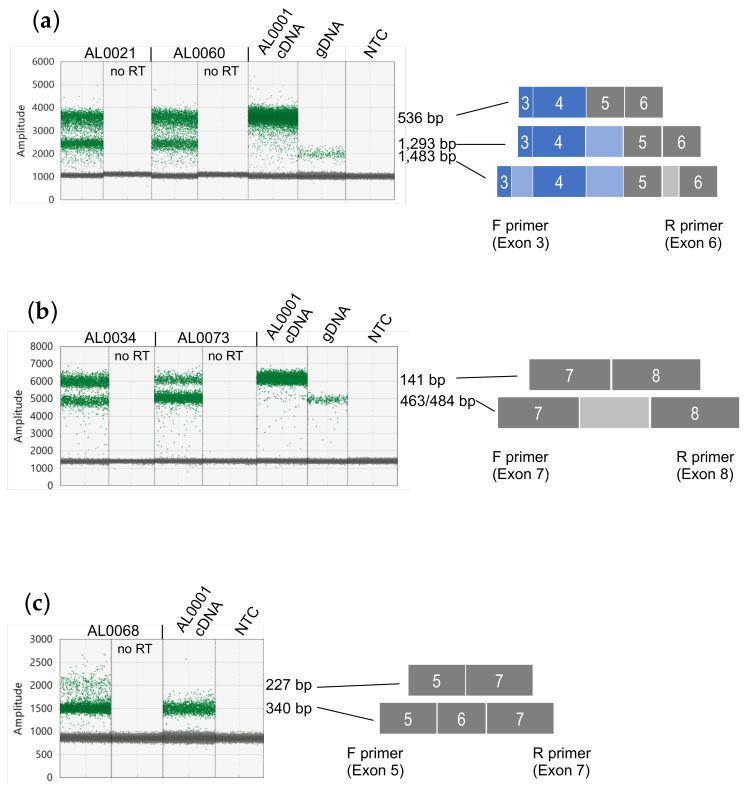
Figure 3.
Validation of TP53 splicing mutations in breast cancer samples. (a) ddPCR assay between TP53 exons 3 and 6 showing amplification of TP53 transcripts. AL0021 and AL0060 have a point mutation in the TP53 intron 4 donor splice site at +1 and +5, respectively. AL0001 has no splicing mutations and was used as a control to show the fluorescence amplitude of a correctly spliced TP53 RNA (536 bp), whereas those TP53 RNAs retaining intron 4 have an amplicon size of 1293 bp. Genomic DNA (gDNA) was used as a control template to show an amplicon with retention of introns 3, 4 and 5. (b) ddPCR assay between TP53 exons 7 and 8 showing amplification of TP53 transcripts. AL0034 and AL0073 have a 21 bp deletion spanning the TP53 intron 7 donor splice site or a point mutation at +1 in the TP53 intron 7 donor splice site, respectively. AL0001 has no splicing mutations and was used as a control to show the fluorescence amplitude of a correctly spliced TP53 RNA (141 bp), whereas those TP53 RNAs retaining intron 7 have an amplicon size of 463 or 484 bp for AL0034 and AL0073, respectively. Genomic DNA (gDNA) was used as a control template to show an amplicon with retention of intron 7 (484 bp). (c) ddPCR assay between TP53 exons 5 and 7 showing amplification of TP53 transcripts. AL0068 has a point mutation at −1 in the TP53 intron 5 acceptor splice site. AL0001 has no splicing mutations and was used as a control to show the fluorescence amplitude of a correctly spliced TP53 RNA (340 bp), whereas those TP53 RNAs skipping exon 6 have an amplicon size of 227 bp. ddPCR results shown are “1-D” plots, with green dots representing droplets where PCR products have been amplified and grey dots represent no amplified PCR product. The fluorescence amplitude (Amplitude) on the y-axes is indicative of amplicon size/s within each assay.