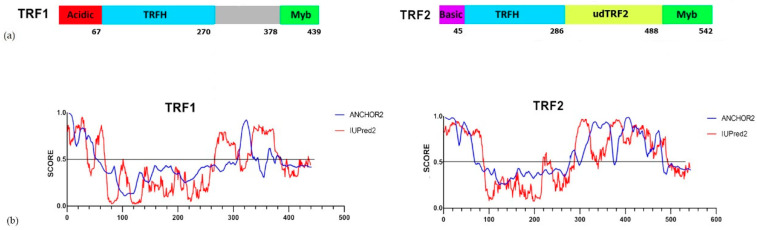
Figure 1.
(a) Comparison of domain structures of TRF1 and TRF2. Basic, basic domain; Acidic, acidic domain; TRFH, TRF homology domain; Myb, DNA-binding Myb-domain. Numerals indicate the number of amino acid residues. (b) Prediction of the secondary structure dynamics of TRF1 and TRF2. IUPred2 (red) and ANCHOR2 (blue) scores are shown. The X-axis represents the amino acid number indicating its position in the sequence; the Y-axis represents the IUPred2 (red) scores that characterize the disordered tendency at each indicated position along the sequence; residues with a predicted score above 0.5 are considered disordered; ANCHOR2 (blue) scores characterize the probability of each residue being part of a binding region. The udTRF2 region of TRF2 is the intrinsically disordered region (IDR) that separates ordered TRFH and Myb domains. TRF1 also has an IDR between TRFH and Myb domains, but it is about half of udTRF2 and contains fewer residues, that could potentially be part of a binding region.