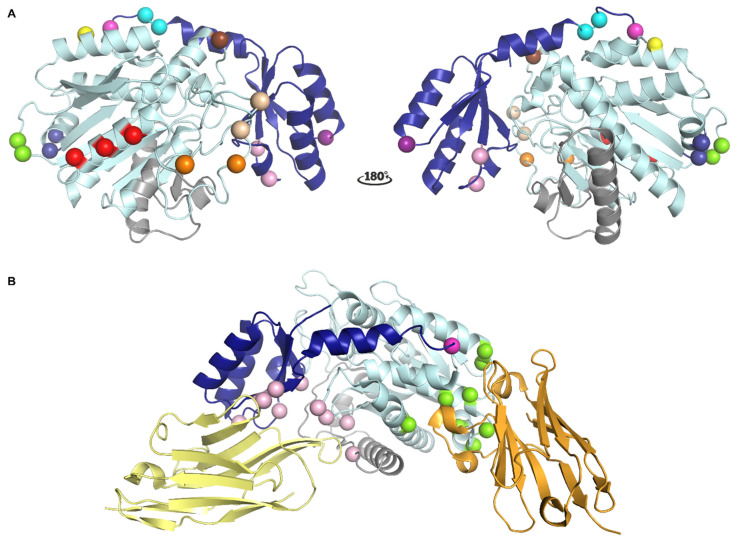
Figure 2.
Localization of different epitopes in the structure of TAFI. (A) Localization of the epitopes of monoclonal antibodies (mAbs) that interfere with TAFI or TAFIa as determined by mutagenesis studies. The activation peptide (AP) and the catalytic moiety are colored in dark and light blue, respectively. The dynamic flap region is colored in grey. Major determinants of the epitopes are indicated as spheres. The cleavage site at Arg92 is indicated as a magenta sphere. Epitope residues for MA-RT36A3F5 and MA-RT13B2 are indicated by dark blue and light orange spheres, respectively. Epitope residues for MA-RT30D8 and MA-RT82F12 are indicated by the light and dark orange spheres. Epitope residues for nanobody VHH-mTAFI-i49, which destabilizes the TAFI proenzyme, are indicated by the brown and light orange sphere indicated by an asterisk. The epitope residue identified for MA-TCK27A4, which interferes with all modes of TAFI activation, is indicated by a yellow sphere. The epitope residue for MA-T12D11, which selectively inhibits T/TM-mediated TAFI activation, is indicated by a purple sphere. Epitope residues for MA-T94H3 and MA-T1C10, which interfere with both T/TM- and plasmin-mediated TAFI activation, are indicated by pink spheres. Epitope residues for MA-TCK22G2, which interferes with plasmin- and thrombin-mediated TAFI activation, are represented by green spheres. Epitope residues for MA-TCK11A9 and MA-TCK26D6, which mainly inhibit plasmin-mediated TAFI activation, are indicated by red and cyan spheres, respectively. Panel A was generated using the structure of intact human TAFI (PDB ID 3D66). (B) Cartoon representation of the crystal structure of TAFI in complex with nanobodies VHH-i83 (yellow) and VHH-a204 (orange) (PDB ID 5HVH). Residues that are engaged in polar interactions with VHH-i83 and VHH-a204 are indicated by pink and green spheres, respectively. The cleavage site at Arg12 is indicated as a magenta sphere.