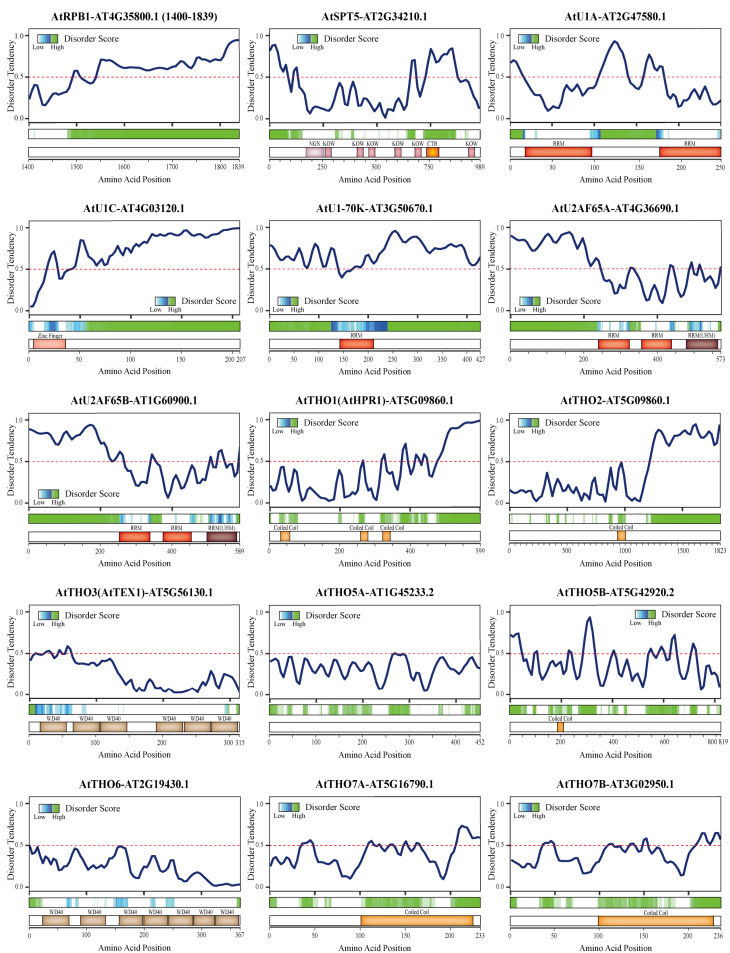
Figure 2.
Disorder tendency predictions for proteins involved in the coupling of transcription and RNA processing in plants. All protein sequences were downloaded from TAIR (https://www.arabidopsis.org/ (accessed on 6 November 2020)). (For each figure, top panel) Protein disorder tendency curve. The disorder tendency score of each amino acid was predicted using IUPred2A [134] and subsequently fit to a smooth curve using the R ggplot2 package. The predicted scores were between 0 and 1, with a score above 0.5 (dashed line) indicating disorder. (For each figure, middle panel) Prediction of disordered regions using D2P2 [135]. (For each figure, bottom panel) A domain map of the proteins in Arabidopsis based on previous studies [81,88] and the protein databases UniPro [136], Pfam [137], and SMART [138]. The disorder predictions for full-length proteins, except for AtRPB1 (C-terminal domain), are displayed. Abbreviations: WD40, tryptophan-aspartic acid motif repeats; RRM, RNA recognition motif; UHM, U2AF homology motif; NGN, NusG N-terminal domain; KOW, Kyrpides–Ouzounis–Woese motif; CTR, C-terminal repeat region.