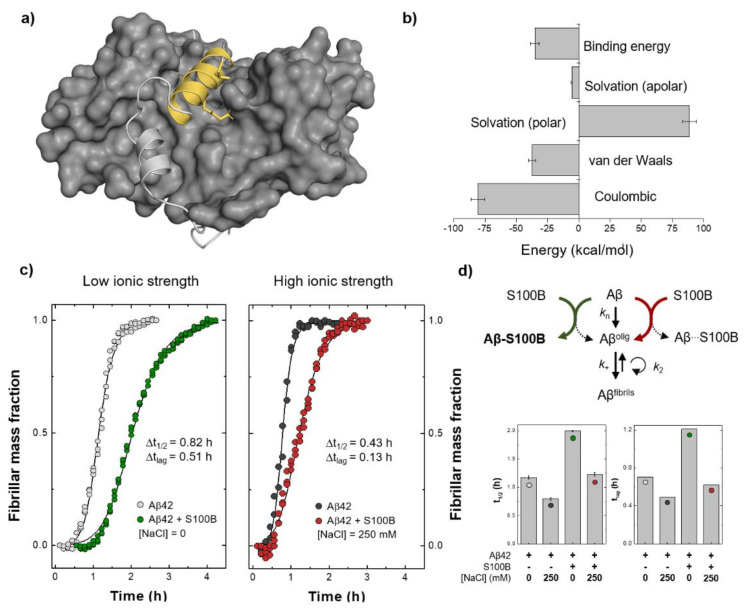
Figure 4.
Estimation of the binding energy for the Aβ:S100B complex and experimental evidence supporting the relevance of coulombic interactions for Aβ:S100B complex stabilization. (a) Structural representation of the Aβ:S100B complex (depicted as light grey cartoon with Aβ25–35 in yellow, on the protein grey surface) highlighting the contributions of Lys28 and Ile31 residues (marked with sticks). (b) Molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) binding energy calculated for the Aβ25–35:S100B complex as the sum of all energetic terms involved over the equilibrated segments of the simulations and with the error bars calculated from the standard error of the mean between replicates. (c) Aggregation of 5 µM Aβ42 at 37 °C with or without 25 µM Ca2+-S100B under low (left) and high (right) ionic strength conditions. (d) Binding impairment between S100B and monomeric Aβ42 at high ionic strength accounts for the partial depletion of S100B inhibitory activity, for example, over the mechanism of primary nucleation, which is exclusively dependent on monomeric Aβ42 concentration (top). Half-time (t1/2) and lag time (tlag) values of Aβ42 aggregation in all tested conditions (error bars represent standard deviation, n = 3) (bottom).