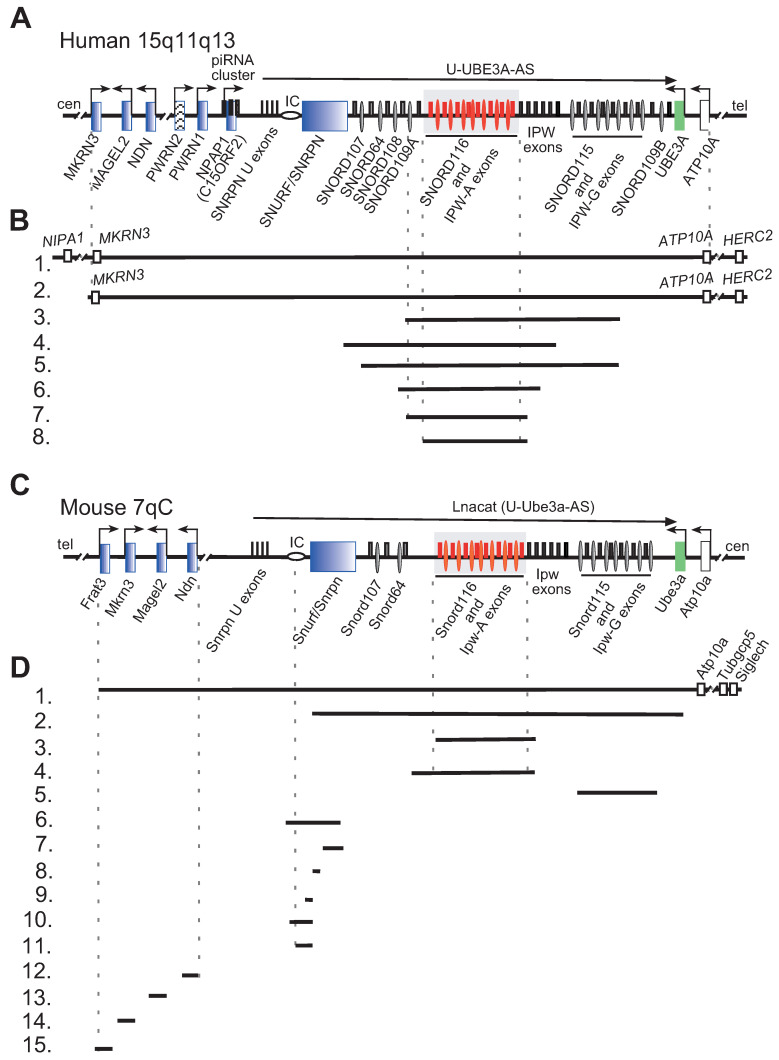
Figure 1.
Organization of human and mouse PWS loci, deletions in human and PWS mouse models are indicated. (A) Schematic representation of the human PWS locus on chromosome 15q11-q13. Blue rectangles denote paternally imprinted protein coding genes. Thin ovals show snoRNA gene locations; the imprinting center (IC) is denoted by a horizontal oval. Thin rectangles above the midline depict non-protein coding exons. SNORD116 and IPW-A exons are displayed in red and further highlighted by a grey rectangle. Arrows indicate promoters and the direction of transcription. The long arrow on top shows the putative U-UBE3A antisense transcript harboring the SNORD116 and SNORD115 clusters. Centromere and telomere regions are indicated as cen and tel. (B) Schematic representation of PWS chromosomal deletions. Lines 1 and 2 indicate the common 5–6 Mb PWS deletion [2]. Lines 3–8 represent the characterized PWS cases with microdeletion in the Snord116 array. Line 3.—[34], 4.—[35], 5.—[36], 6.—[37], 7.—[38], 8.—[39] (C) Schematic representation of the mouse PWS-locus on chromosome 7qC (symbols as above). (D) Schematic representation of available mouse models in PWS research. (1.) The largest chromosomal deletion that eliminates the PWS/AS region and a large portion of non-imprinted genes [40]. (2.) Deletion of the mouse PWS-locus span from the Snurf/Snrpn to Ube3a genes [41]. (3.) Deletion of the PWS critical region (~300 kb) (PWScr) comprising of Snord116 and IPW-A gene arrays [42]. (4.) The Snord116del mouse model eliminating a larger ~350 kb PWScr genomic region [43]. Note, that the genomic assembly of PWScr is not completed, a gap of ~50 kb inside the Snord116 cluster might increase the snoRNA gene copy number and overall size of deletion (UCSC, GRCm39/mm39 chr7:59457067- 59507068). (5.) Deletion of the Snord115 gene cluster [44] (6.–11.) Deletions within the Snurf/Snrpn and PWS IC center (Details in Figure 2). (6.) 35 kb deletion of IC center and Snurf/Snrpn exons 1–6 [45]. (7.) Deletion of Snurf/Snrpn exon 6 including parts of exons 5 and 7 [45]. (8.) Deletion of Snurf/Snprn exon 2 [41]. (9.–11.) Elimination of Snurf/Snrpn exon 1 and upstream genomic region: 0.9 kb (8), 4.8 kb (9) and 6 kb (10) deletions, respectively [46,47]. (12.–15.) Deletion of protein coding genes within the PWS-locus: (12.) Ndn [48,49,50,51]; (13.) Magel2 [52,53]; (14.) Mkrn3 [54]; (15.) Frat3 [30].