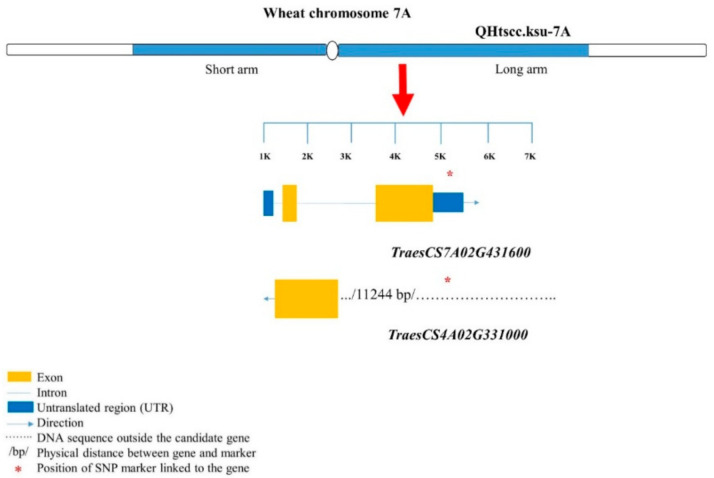
Figure 4.
Structures of candidate genes in the QHtscc.ksu-7A genomic region (GR). The genes were identified based on the 90K wheat microarray assessment of near-isogenic lines with contrasting root traits. SNP markers are located in different positions of the genes. Structural information of the genes and SNPs was from a wheat genome database (https://urgi.versailles.inra.fr/jbrowseiwgsc/gmod_jbrowse/, accessed on 2 March 2021). The physical positions of the linking markers of the QTL were found from the original mapping study using parental cultivar; therefore, in the reference genome (Chinese Spring), the QTL extends in both the long and short arm of chromosome 7A, which is unusual. Chromosomal fragment translocation might be the possible reason for the difference between the reference genome and the parental cultivars used in the original mapping study [25]. The measuring bar indicates the DNA length in kilo base pairs (kb).