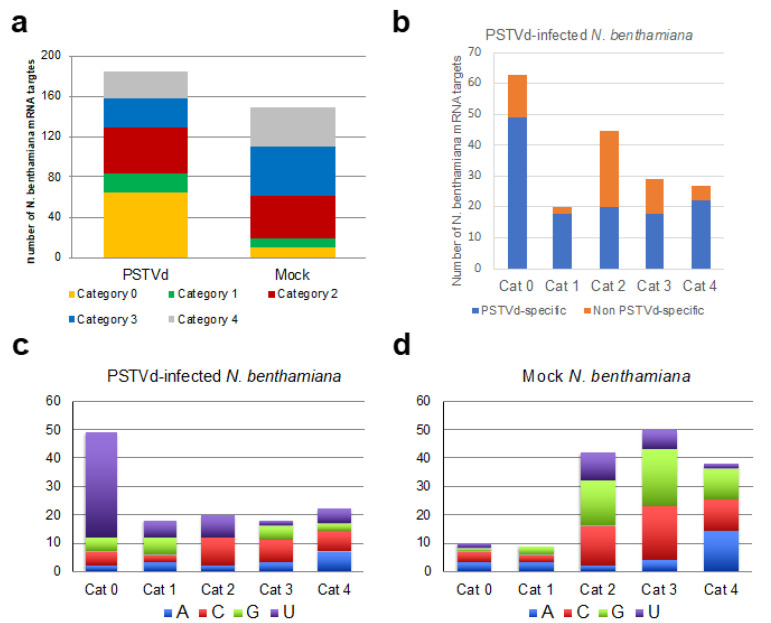
Figure 6.
(a) Total number and quality categorization (from 0 to 4, where 0 represents the strongest evidence for true cleavage products) of mRNA targets of PSTVd-sRNA and non-PSTVd specific (false positive) mRNAs targets identified by degradome analysis in PSTVd-infected and mock-inoculated N. benthamiana, respectively; (b) number of specific and non- PSTVd specific mRNA targets in PSTVd-infected N. benthamiana plant in each degradome category. The PSTVd-specific mRNA targets were those not identified in any of the two mock-inoculated tomato replicates; (c) analysis of the 5′ terminal nucleotide of PSTVd-sRNAs involved in the cleavage of PSTVd-specific targets according to the degradome category in PSTVd-infected N. benthamiana, the 5′ terminal nucleotide of PSTVd-sRNAs involved in host mRNA cleavages classified into the category 0 were mainly U. (d) When the same analysis was performed considering the PSTVd-sRNAs targeting the false (non-PSTVd-specific) targets identified in the degradome sequencing from mock-inoculated samples, no bias was observed in the 5′ terminal nucleotide of PSTVd-sRNAs targeting mRNAs of category 0, suggesting that they were not targeted by AGO1.