Table 1.
Bona fide mRNA targets of PSTVd-sRNA in tomato identified by degradome analysis.
| mRNA Target | Degradome Results | PSTVd-sRNAs | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. Thaliana Homolog | Alignment Score | mef (Kcal/mol) | PSTVd-sRNA:mRNA Hybrid | Cat | Cleavage Position | p-Value T3 | Number of Tags T3 | Normalized Weighted Number of Tags T3 | p-Value T4 | Number of Tags T4 | Normalized Weighted Number of Tags T4 | 5′Position | Polarity | PSTVd Variants * | Reads T3 | RPM T3 | Reads T4 | RPM T4 | |
| Solyc09g083420.2.1 | AT1G56290.1 (CWF19) | 2.5 | −37.4 |

|
0 | 1587 | 0.00 | 74 | 1.93 | 0.00 | 149 | 2.62 | 178 | (+) | Nb, Int, Mild, RG1, Lethal | 935 | 25.22 | 1330 | 35.89 |
| Solyc05g007450.2.1 | AT1G26840.1 (ORC6) | 3.0 | −29.6 |

|
0 | 645 | 0.00 | 113 | 2.95 | 0.00 | 259 | 4.55 | 179 | (+) | Nb, Int, mild, RG1, Lethal | 2037 | 54.97 | 2931 | 79.1 |
| Solyc11g012270.1.1 | AT3G07880.1 (SCN1) | 3.5 | −31.0 |
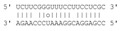
|
0 | 451 | 0.01 | 8 | 0.21 | 0.00 | 13 | 0.23 | 192 | (+) | Nb, Int, RG1, Lethal | 409 | 11.03 | 488 | 13.17 |
| Solyc03g045050.2.1 | AT1G67230.1 (LINC1) | 3.5 | −34.4 |
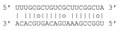
|
0 | 3520 | 0.00 | 196 | 5.12 | 0.00 | 316 | 5.55 | 238 | (+) | Nb, Int, Mild, RG1, Lethal | 2096 | 56.56 | 5535 | 149.4 |
| Solyc05g024290.2.1 | AT5G13160.1 (PBS1) | 3.0 | −29.6 |

|
0 | 1542 | 0.00 | 56 | 1.46 | 0.01 | 163 | 2.86 | 290 | (+) | Nb, Int, RG1, Lethal | 138 | 3.72 | 542 | 14.63 |
| Solyc12g007200.1.1 | AT4G28980.1 (CDKF;1) | 3.5 | −33.7 |

|
0 | 761 | 0.01 | 28 | 0.73 | 0.00 | 142 | 2.49 | 8 | (+) | Nb, Int, mild, RG1, Lethal | 277 | 7.47 | 354 | 9.553 |
| Solyc04g079940.2.1 | AT2G24430.1 (ANAC038) | 3.0 | −25.5 |

|
0 | 1339 | 0.00 | 33 | 0.86 | 0.02 | 38 | 0.67 | 70 | (−) | Nb, Int, mild, RG1, Lethal | 18 | 0.49 | 85 | 2.29 |
| Solyc07g063100.2.1 | AT4G27500.1 (PPI1) | 3.0 | −28.4 |

|
0 | 1675 | 0,00 | 637 | 16.65 | 0.00 | 2327 | 40.87 | 288 | (−) | Nb, Int, mild, RG1, Lethal | 319 | 8.61 | 458 | 12.36 |
