Figure 4.
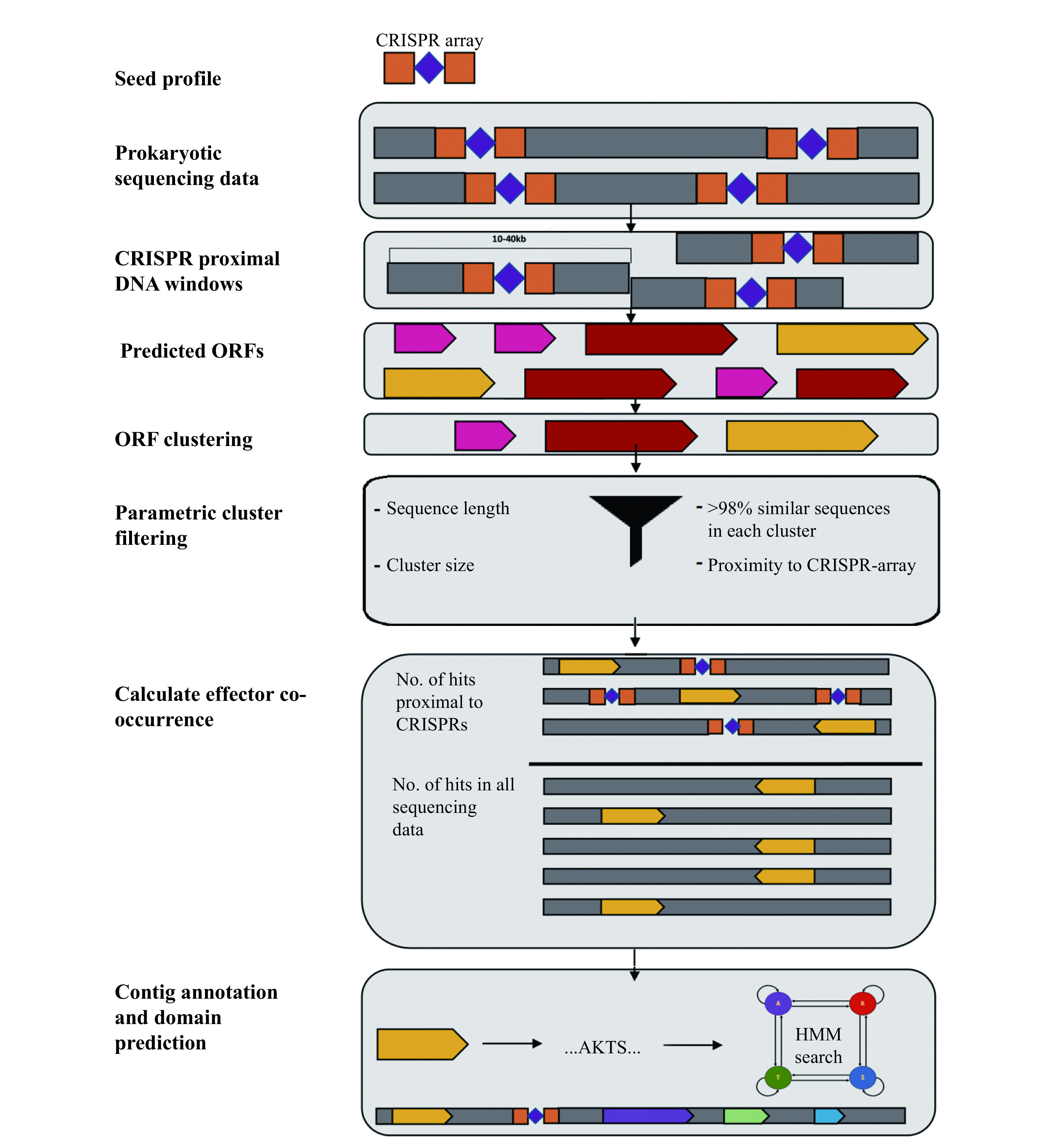
Overview of the computational pipeline used to uncover CRISPR associated proteins.
Proteins were recovered from sequence assemblies by extracting the sequences upstream and downstream of CRISPR arrays and determining whether open reading frames predicted from these sequences co-occurred with CRISPR arrays. A co-occurrence score was calculated as the number of instances of a particular protein homolog encoded within a pre-defined number of bases from a CRISPR-array divided by the total number of instances of the same homolog in all sequencing data used as the starting input to the pipeline.
