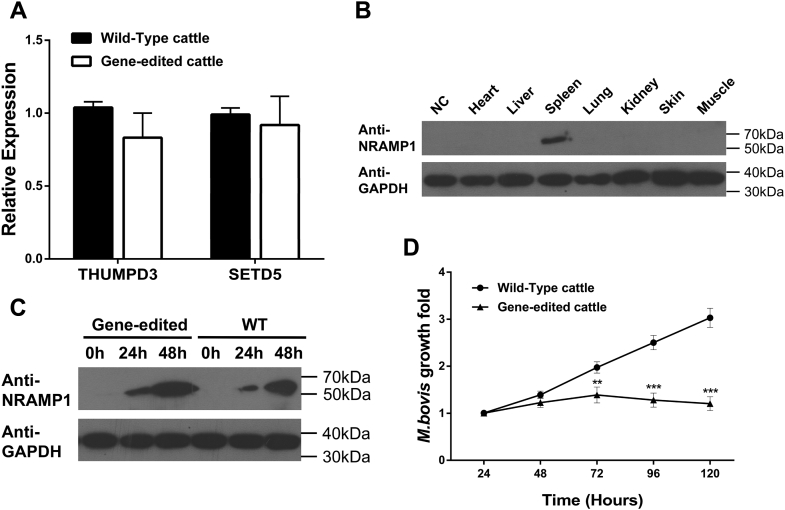
Figure 5.
Assessment of the increased resistance of gene-edited cattle to tuberculosis.A, the relative expression levels of the nearby endogenous genes at the ROSA26 locus by qPCR. B, western blot analyses to detect NRAMP1 expression using the goat anti-rabbit NRAMP1 polyclonal antibody. The organs were obtained from a pool of dead gene-edited cattle. Donor cells were used for negative control (NC). C, the expression of NRAMP1 was highly activated in the gene-edited cattle following M. bovis infection. All the samples were mixed MDMs that were isolated from the blood of gene-edited cattle as a pool. “WT” represents wild-type cattle. D, multiplication of M. bovis in MDMs from wild-type cattle or gene-edited cattle in vitro. Data are presented as the mean ± SD of three independent experiments. Student's t-test was used to evaluate the differences. ∗∗p < 0.01; ∗∗∗p < 0.001.