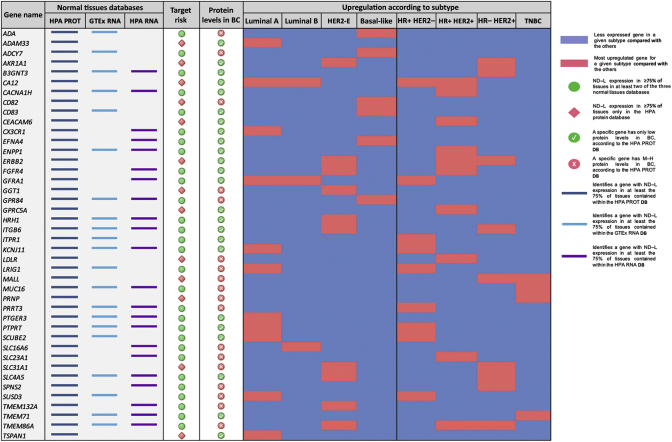
Figure 2.
Weak and strong gene candidates, with toxicity risk category and breast cancer subtype-specific upregulation.
HPA PROT, Human Protein Atlas database of protein expression in normal tissues; GTEx RNA, Genotype-Tissue Expression database of mRNA levels of human genes in normal tissues; HPA RNA, Human Protein Atlas database of mRNA levels of human genes in normal tissues. For each database column, the presence of a colored segment identifies when the corresponding gene presented a not detected and/or low (ND–L) expression in at least 75% of tissues contained within such dataset. The column ‘Target Risk’ identifies the concordance of expression levels in the three normal tissues' databases. A green circle represents an ND–L expression in ≥75% of tissues in at least two of the three databases, while a red rhombus represents an ND–L expression in ≥75% of tissues only in the HPA protein database. The column ‘Protein Levels in BC’ identifies the levels of protein expression for each gene in breast cancer tissue, as per HPA cancer tissue database. Red circles represent genes with only low protein levels, while green circles represent genes with medium/high protein levels. In the ‘Upregulation according to subtype’ section, for each breast cancer subtype the blue color identifies a less expressed gene (database TCGA, result from differential expression analysis) and the red color represents an upregulated gene for a subgroup compared with the others.