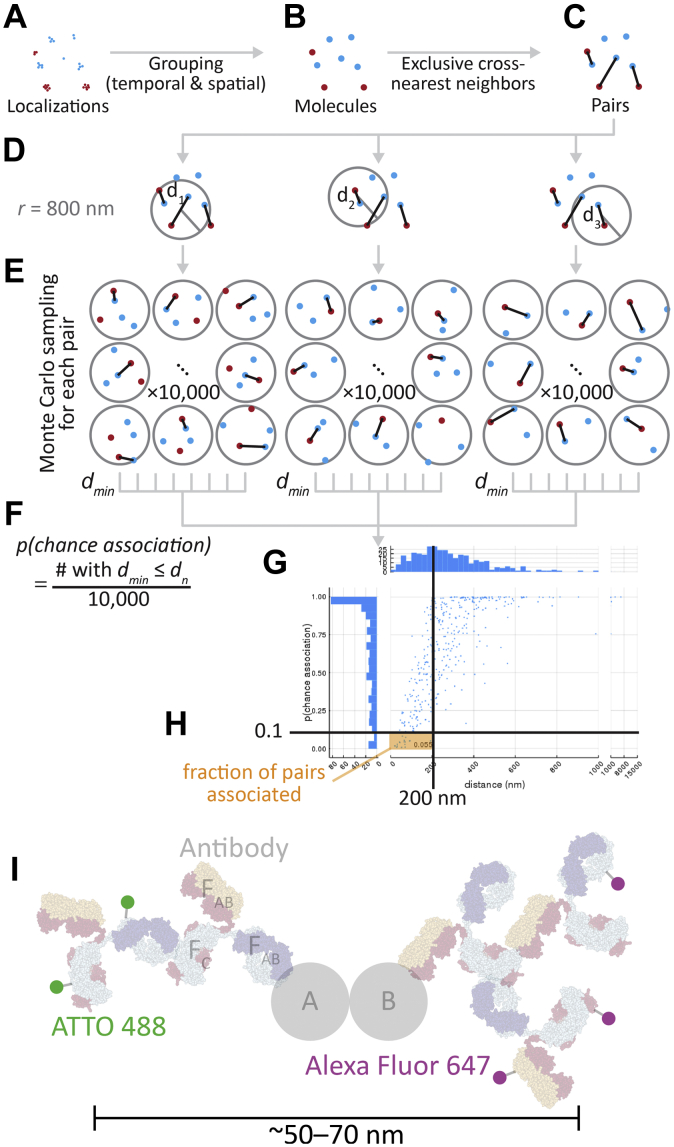
Figure 1.
Cross-nearest neighbor/Monte Carlo method to estimate the fraction of molecules associated. Scattered localizations (A) were grouped over time and space to into “molecules” which have a position that is the average of their component localizations (B; see Fig. S5). These molecules were exclusively paired to their cross-nearest neighbors (C). For each pair (D), 10,000 permutations of the molecules within radius r (800 nm) of the centroid of the pair were generated, and the closest intermolecular distance was measured (E). The fraction of events less than the pair’s distance was the probability of chance association (p(chance association)) (F). These values were accumulated across the whole cell (plotted in G), and the fraction of pairs with a probability of chance association <0.1 and within a physically possible binding distance (<200 nm), the fraction associated, was calculated (H). I, The physical arrangement of a bound pair and antibody stack. Typical immunofluorescence uses expensive, target-specific primary antibodies, and cheap secondary antibodies conjugated to a fluorophore like ATTO 488 and Alexa Fluor 647. For orientation, one antibody is labeled for its constant domain (FC) and two antigen binding domains (FAB). The physical size and arrangement mean that ∼50 to 70 nm may separate signal from the two fluorophores when detecting a binding interaction between proteins A and B, and multiple fluorophores can produce signal spread over tens of nanometers. Antibody graphic was created using NGL Viewer (56) from RCSB PDB 1IGT.