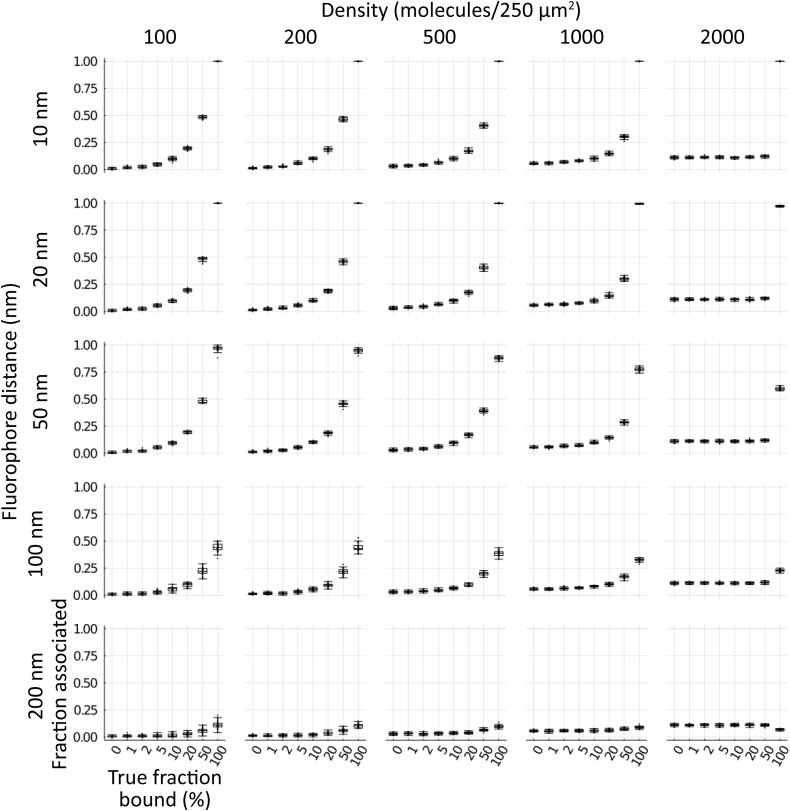
Figure 3.
Density and pair distance influence fraction associated. Simulated cells of 100, 200, 500, 1000, and 2000 molecules of each type (columns left to right) within a 250 μm2 circle with 10, 20, 50, 100, and 200 nm separation between pairs (rows top to bottom) for cells with 0, 1, 2, 5, 10, 20, 50, or 100% binding were generated (n = 30 each condition). Molecules for each simulated cell were run through the cross-nearest neighbor/Monte Carlo algorithm and median distance between pairs measured. Boxes indicate median ± upper and lower quartile; whiskers indicate the range excluding outliers.