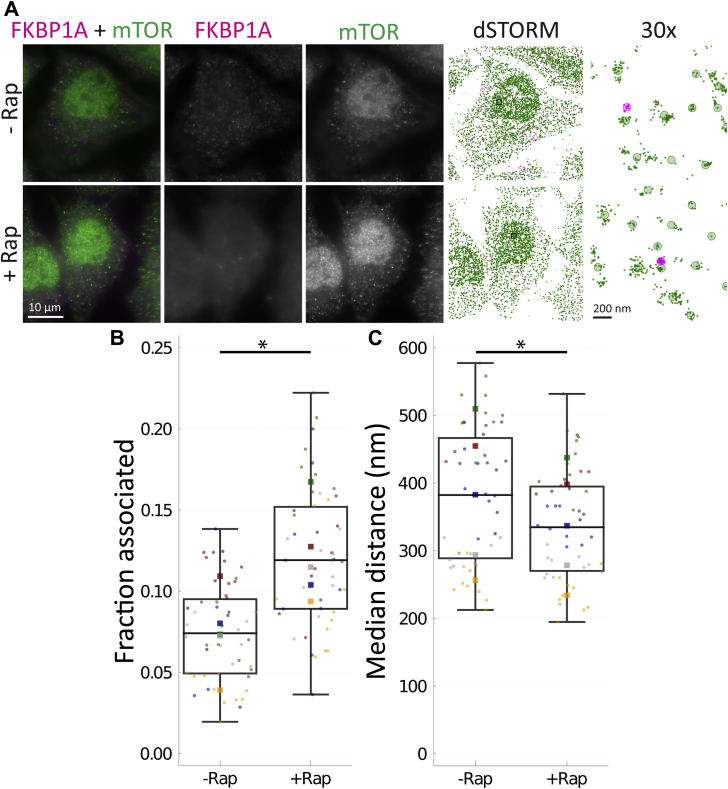
Figure 4.
Method detects rapamycin-induced FKBP1A association with mTOR. U2OS-MEG3 cells were treated for 24 h with or without 10 ng/ml rapamycin. A, cells were stained for mTOR with a secondary antibody conjugated to ATTO 488 (green) and for FKBP1A with a secondary antibody conjugated to Alexa Fluor 647 (magenta). From left to right: Merged image; RNA channel; p53 channel; dSTORM localization map; 30x inset of dSTORM localizations in the black box, with shaded circles indicating “molecules”. Scale bars are 10 μm, or 200 nm (right column). B, fraction of pairs associated, as defined by a probability of chance association <0.1 (i.e., correction for local density) and distance <200 nm (upper limit for binding distance, accounting for error). C, median distance between pairs for each cell (nm). Boxes indicate median ± upper and lower quartile; whiskers indicate the range excluding outliers. Data points are colored by replicate. For each condition, single molecule localizations were collected from 10 randomly chosen cells in five separate experiments. Means for each replicate are indicated by same-colored squares. ∗ indicates p < 0.05 by ANOVA. dSTORM, direct stochastic optical reconstruction microscopy.