Extended Data Fig. 4. Base editing of the BCL11A and NF-Y motif.
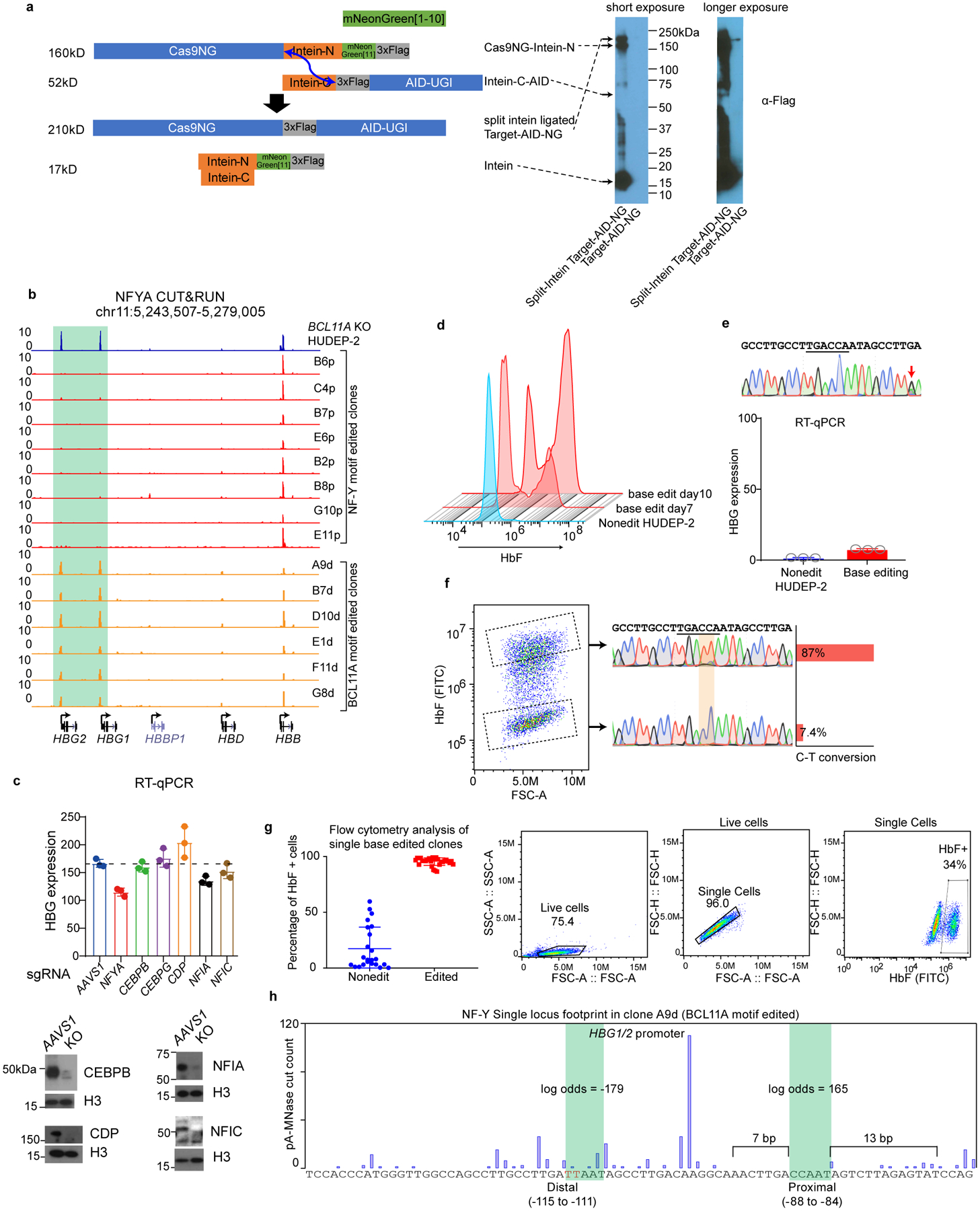
(a) Left, split-intein mediated ligation of Cas9NG-Intein-N and Intein-C-AID, producing full-length Target-AID-NG. Blue arrow indicates the ligation sites. Right, immunoblot validating the expression of each component and the ligation products. The ligation is incomplete, but the level of ligated product is much higher than the original vector (cropped).
(b) NFYA binding at the γ-promoters diminished in all the NF-Y motif-edited clones (red), and increased in all the BCL11A motif-edited clones (orange), as revealed by NFYA CUT&RUN. NF-Y motif editing was carried out in BCL11A KO HUDEP-2 cells while BCL11A motif editing was carried out in wild-type HUDEP-2 cells.
(c) Upper, RT-qPCR analysis of γ-globin expression after acute depletion of C/EBPβ, C/EBPγ, CDP, NFIA and NFIC. Lower, immunoblot validating protein depletion (cropped). BCL11A KO HUDEP-2 cells were differentiated for 3 days after nucleofection. The result is shown as mean (SD) of three technical replicates.
(d) Flow cytometry analysis of HbF levels for BCL11A base-edited clones at day 7 and 10. Longer editing resulted in higher base editing rate (Figure 3e) and higher percentage of HbF positive cells.
(e) A control base editing experiment in which a nucleotide 9 bp away from the BCL11A motif was edited. Sanger sequencing confirmed C-T conversion.
(f) Left, FACS of BCL11A motif base-edited bulk cells into high and low HbF populations. The C-T conversion rate of BCL11A motif in each population was measured by Sanger sequencing and quantified with TIDER. HbF high cells show 87% conversion and HbF low cells show only 7.4% conversion.
(g) Left, flow cytometry analysis of HbF level in individual clones derived from BCL11A motif base editing. Data is showed as mean (SD) of multiple independent clones. Nonedit: n=23, base edited: n=30. Right, gating strategy.
(h) Single locus footprint of NF-Y at the γ-promoters in clone A9d, a BCL11A motif-edited clone.
