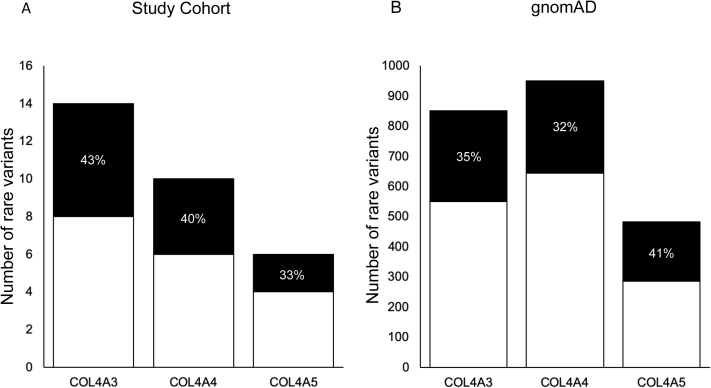
Figure 1.
Number of rare missense variants predicted to be pathogenic in: (A) the focal segmental glomerulosclerosis (FSGS) study cohort and (B) Genome Aggregation Database (gnomAD). For rare missense COL4A3, COL4A4, and COL4A5 variants in our FSGS cohort, 43% (6/14), 40% (4/10), and 33% (2/6) were predicted to be deleterious by at least 10 of 12 programs, respectively. For rare missense COL4A3, COL4A4, and COL4A5 variants identified in gnomAD, 35% (301/851), 32% (306/949), and 41% (197/483) were predicted to be deleterious by at least 10 of 12 programs, respectively.