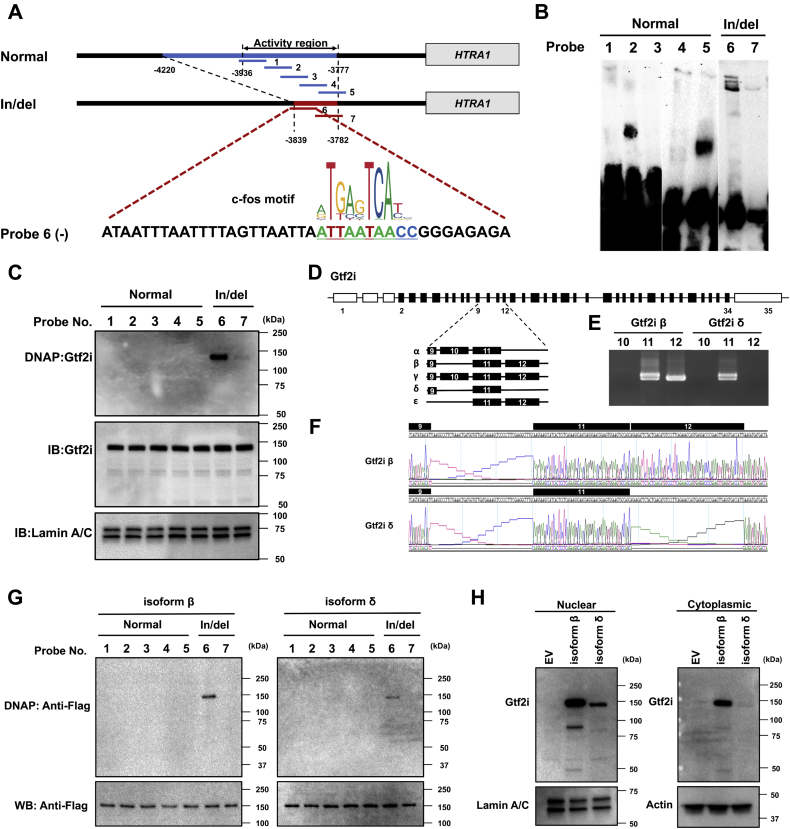
Figure 1.
In/del binding transcription factor protein. A, schematic illustration of the double-stranded DNA probes for EMSA (gel electrophoresis mobility shift assay) in the region upstream of the HTRA1 activity coding region. Linking c-fos transcription factor to in/del-6. Profile of the c-fos transcription factor-binding sequence (ID: MA0099.3 from the JASPAR database). B, EMSA was performed to determine the in/del binding activity protein. In total, 50 μl nuclear protein from 661W cells was incubated with 100 pmol biotin-labeled Double-stranded DNA probes and analyzed on a 7.5% EMSA gel. Bands of interest were cut out and processed for LC-MS/MS analysis. C, Gtf2i-DNA probes binding test. The binding ability between Gtf2i and the in/del region was confirmed by WB. In/del DNA probes 6 and 7 were detected by an anti-Gtf2i antibody. Detection of Lamin A/C was used as an internal control. D, gene structure of Murine Gtf2i. Coding exons are depicted as black boxes and noncoding exons are in blank boxes. Five isoforms signify various alternatively spliced isoforms with exon 9, 10, 11, and 12 indicated. In 661W cells, there are only isoforms β and δ present by TA cloning (E, F). G, Gtf2i β/δ -DNA probes binding test. Both Gtf2i β and δ bind to the in/del-6 probe by WB. H, expression of Gtf2i isoform β and δ in nuclear or cytoplasmic extracts. Vector of Gtf2i isoform β or δ was transfected into 661W cells, followed by WB. Gtf2i isoform β was expressed in both nuclear and cytoplasmic extracts. However, isoform δ was only detected in nuclear but cytoplasmic extracts.