Abstract
In this study, we have investigated the binding mechanism of two FDA-approved drugs (ivermectin and levosalbutamol) with the spike protein of SARs-CoV-2 using three different computational modeling techniques. Molecular docking results predict that ivermectin shows a large binding affinity for spike protein (− 9.0 kcal/mol) compared to levosalbutamol (− 4.1 kcal/mol). Ivermectin binds with LEU492, GLN493, GLY496, and TRY505 residues in the spike protein through hydrogen bonds and levosalbutamol binds with TYR453 and TYR505 residues. Using density functional theory (DFT) studies, we have calculated the binding energies between ivermectin and levosalbutamol with residues in spike protein which favor their binding are − 22.4 kcal/mol and − 21.08 kcal/mol, respectively. The natural bond orbital (NBO) charge analysis has been performed to estimate the amount of charge transfer that occurred by two drugs during interaction with residues. Molecular dynamics (MD) study confirms the stability of spike protein bound with ivermectin through RMSD and RMSF analyses. Three different computer modeling techniques reveal that ivermectin is more stable than levosalbutamol in the active site of spike protein where hACE2 binds. Therefore, ivermectin can be a suitable inhibitor for SARS-CoV-2 to enter into the human cell through hACE2.
Keywords: SARS-CoV-2, COVID-19, hACE2, Docking, Ivermectin, MD simulation
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infests the respiratory tract and causes coronavirus disease 2019 (COVID-19). It is a new member of the coronavirus family which is closely related to SARS-CoV in genetic sequencing [1, 2]. SARS-CoV-19 was first recognized in December 2019 which was globally spread and infected 80,773,033 people including 1,783,619 deaths according to a report of “World Health Organization” (9.32 pm GMT+6, 30 December 2020). The cause of the worldwide outbreak of this virus is the ability to transmit man to man by contact with infected droplets of saliva, cough, or sneeze [3]. Its high infective potential is disrupting the economic and social life of every single people worldwide. It is reported that in 20% of infected people, COVID-19 spreads in the lungs and other organs of the body. But, no drug or vaccine has been approved against COVID-19. A few FDA-approved drugs are being used on COVID-19 patients at the early stage of infection which may cause less severe infection due to reduced entry of COVID-19 into the cell. Ivermectin, an FDA-approved antiparasitic drug, can reduce the replication of SARS-CoV-19 in vitro [4]. Many research groups have proved that SARS-CoV-2 utilizes the homotrimeric spike (S) glycoprotein to enter into the human cell through binding to the functional receptor human angiotensin-converting enzyme 2 (hACE2) [5, 6]. The receptor-binding motif (RBM) in the S protein of SARS-CoV-2 has two regions that form the interface between S protein and hACE2 [7]. In this interface, the existing L455 and Q493 of the SARS-CoV-2 have favorable interaction with hACE2 and K31 and E35 in hACE2 form hydrogen bonds with Q493 to enhance binding S protein [5]. If any drug molecule can inhibit the binding between SARS-CoV-2 and hACE2 by blocking the S protein’s interface, especially Q493 and L455, it will prohibit the SARS-CoV-2 entry into the human cell.
In the present study, we investigated the interaction of ivermectin with the interface region of the spike protein through which it binds with hACE2. We also tried to understand which amino acid residues in the spike protein are favorable to bind with ivermectin and how they interact? The molecular docking was conducted to predict the active site at the S protein interface for ivermectin and their potential binding affinity and interactions. Density functional theory (DFT) was carried out to calculate the binding energy between interacting residues and ivermectin and the amount of charge transfer during an interaction. The molecular dynamics (MD) simulation was performed to understand the stability of spike protein bound with ivermectin through RMSD and RMSF analyses [8]. We considered one more FDA-approved drug levosalbutamol, a β2 adrenergic receptor with a poor antiviral effect [9]. This drug is used for asthma and chronic obstructive pulmonary diseases (COPD) [10].
Computational methodology
Molecular docking
The crystal structure of the spike protein of COVID-19 was recovered from the protein data bank (PDB:6m0j) at 2.45-Å resolution. The water molecules and heteroatoms were removed and polar hydrogen atoms were added to the amino acid residues. The initial structure of ivermectin was obtained from the PubChem database and levosalbutamol was constructed through GaussView. All prepared PDB files of protein and drug molecules were processed to PDBQT format for reckoning energy grid map.
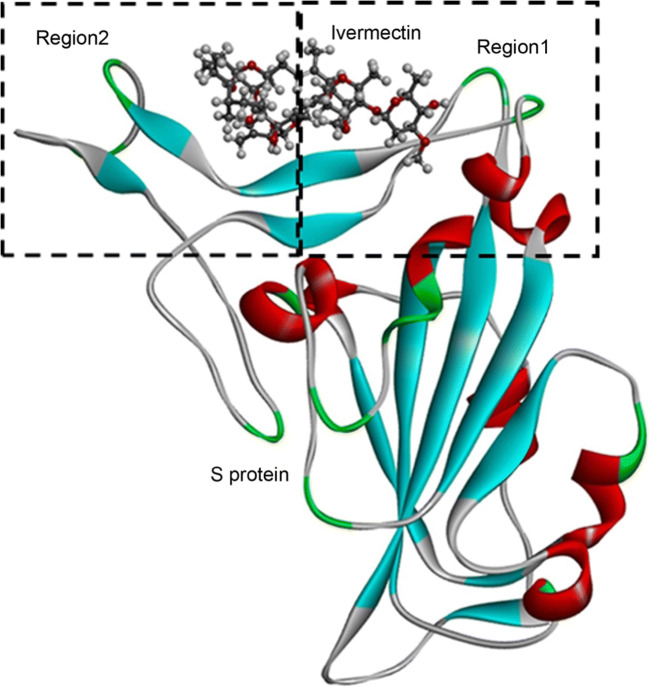
The binding affinities of these two drugs with S protein were elucidated by AutoDock Vina. The docking grid was set around the region 1 and region 2 of spike protein (Fig. 1) with center X = − 37.33, Y = 31.93, and Z = 2.68 and the dimension were X = 29.50, Y = 44.97, and Z = 19.33. The docking process was repeated 10 times and the mean affinity score was considered as binding affinity. In every docking process, two drug molecules were kept flexible. To visualize the docking results PyMol and Discovery Studio Visualizer were utilized.
Fig. 1.
The model of the spike protein bound with ivermectin. Dotted square area right (left) represents the region1 (region 2) in the spike protein of the SARS-CoV-2
DFT calculations
Using docking results, all amino acid residues of the spike protein were selected out which favored two drugs to bind. The coordinates of selected amino acid residues of S protein and drug molecule were considered as an initial configuration to calculate binding energies. Equilibrium structures with local minima in potential energy profiles were computed using density functional theory (DFT) with Becke three-parameters exchange functional [11] and the Lee-Yang-Parr functional [12] (B3LYP) with the all-electron 6-31g(d) basis set. The vibrational frequencies were calculated and no imaginary frequencies were found for the optimized structures. To get binding energies, drug molecules and all selected amino acid residues with frozen backbone atoms were optimized, separately. The binding energy was calculated using Eq. (1) [13]:
| 1 |
where EBE is the binding energy between selected amino acid residues and drug molecules. Eres − lig is the total energy of residues-drug complex after binding. Eres and Elig are total energies of residues and drug molecules, respectively. All calculations are carried out at the gas phase with the Gaussin16 program [14].
Molecular dynamics
Molecular dynamic (MD) simulations were performed to the conformations of S protein bound with ivermectin and levosalbutamol with the highest docking score, separately. Prior to MD simulation, the complexes were immersed in a cubic periodic box and solvated in 19,533 simple point charge (SPC) water molecules during the topology process. Two chlorine ions were added to neutralize the system. 0-nm distance from the box edge and was subjected to energy minimization using 5000-step steepest descent minimization. A 100 ps NVT followed by a 100 ps NPT ensemble at 300 K and 1.0 bar were performed using Berendsen thermostat and barostat[15] to equilibrate water molecules around the complex. The particle mesh Ewald (PME) [16] algorithm was used to consider the long-range Coulomb interactions. For van der Waals interactions, Lennard-Jones potentials were used. A cutoff of 1.2 nm was applied for considering both long- and short-range interactions. The LINCS algorithm was applied to constrain all bonds [17]. A 100-ns-long MD simulation was performed in the NVT ensemble at 300 K with a time step of 2.0-fs and 10.0-ps interval final coordinates were saved. MD simulation was performed using the Gromcas 5.1.4 [18] and force field Charmm36 [19].
Results and discussions
Molecular docking
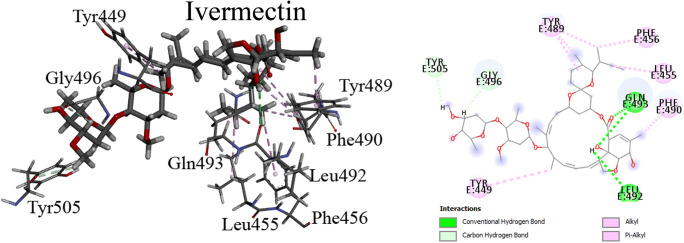
The highest binding affinity of ivermectin and levosalbutamol at the interface of S protein was calculated − 9.0 and − 4.1 kcal/mol, respectively. Figure 2 shows the binding pocket of S protein with ivermectin. Here, residues GLN493 and LEU492 are involved in hydrogen bond (HB) formation with ivermectin. The hydrogen atom of the NH2 group of GLN493 forms two HBs with the oxygen atom of -CO and oxygen atom of –OH groups in ivermectin and the oxygen atom of the –CO group in LEU492 forms one HB with the hydrogen atom of –OH of ivermectin. The HB plays a vital role in stabilizing the protein-ligand complex. Besides these, oxygen atom of –CO groups in TYR505 and GLY496 form HBs with the hydrogen atom of the alkyl group in ivermectin. GLU484, PHE497, ASN501, GLN498, TYR453, SER494, AGR403, and TYR495 interact with ivermectin through van der Waals interaction. Another stacking interaction between aromatic rings of residues (TYR449, TYR489, PHE456, LEU455 and PHE490) and the alkyl group of ivermectin plays a significant role to strengthen the binding of ivermectin with S protein, including hydrogen bonds and van der Waals interaction. Among all interacting residues, GLN493 and LEU455 residues which have favorable interactions with hACE2 can bind with ivermectin. Therefore, before the interaction between S protein and hACE2, it can block the S protein’s binding pocket, which may inhibit SARS-CoV-2 binding with hACE2.
Fig. 2.
3D view of ivermectin with residues inside the active site of spike protein (left). Right figure shows the 2D representation with types of interactions
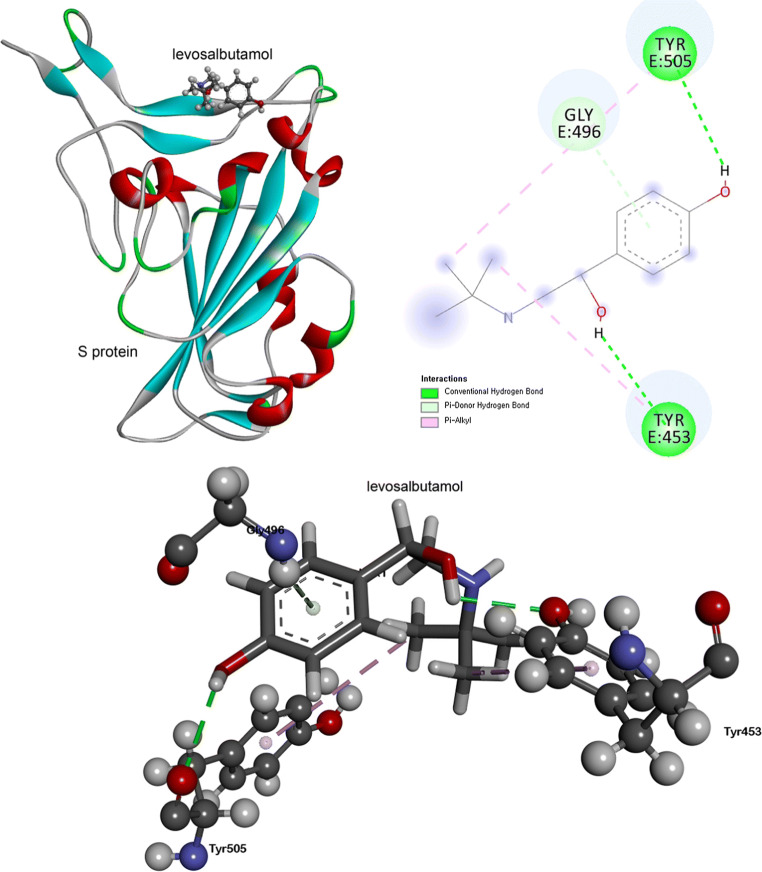
Figure 3 shows the interaction between S protein and levosalbutamol. TYR453 and TYR505 are involved to form two HBs with levosalbutamol. The oxygen atoms of the –OH and –CO group in TYR453 and TYR505 form HBs with hydrogen atoms of –OH groups in levosalbutamol, respectively. Another stacking interaction is found between aromatic rings of residues (TYR453 and TYR505) and alkyl carbon of levosalbutamol. GLY496 residue interacts with levosalbutamol through the pi-donor hydrogen bond.
Fig. 3.
The top figure (left) shows the interaction between levosalbutamol and spike protein after molecular docking. Right (top) and bottom figures show the 2D and 3D representations of interacting residues of spike protein and levosalbutamol with types of interactions, respectively
DFT calculations
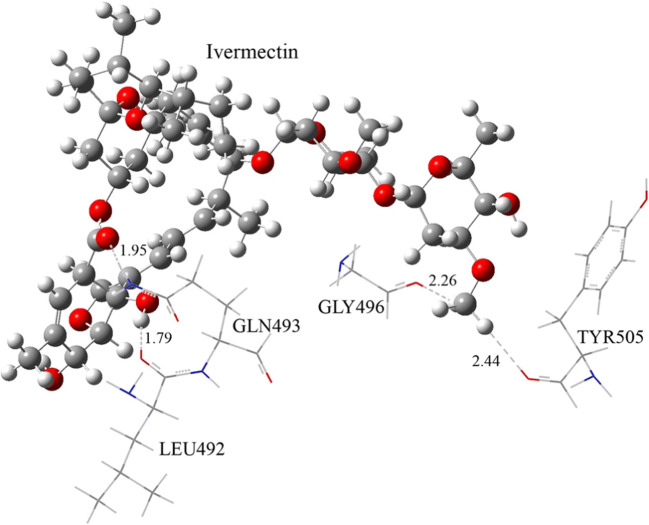
The initial geometries of ivermectin and levosalbutamol were optimized separately at the abovementioned level of theory. We considered only those amino acid residues of S protein that interact with either ivermectin or levosalbutamol through hydrogen bonds to reduce simulation time. Figure 4 shows the optimized configuration of four residues (LEU492, GLN493, GLY496, and TRY505), which interact with ivermectin through hydrogen bonds. The calculated binding energy of these four residues with ivermectin is − 22.4 kcal/mol. Oxygen and hydrogen atoms of –CO and –OH groups of ivermectin form hydrogen bonds with –NH group of GLN493 and –CO group of LEU492, respectively. Alkyl hydrogens of ivermectin form two hydrogen bonds with GLY496 and TYR505. Hydrogen bonds (–CO- -NH-) and (–OH- -OC-) are shorter in distance compared to those form with alkyl hydrogens of ivermectin. The hydrogen bond distances are shown in Fig. 4.
Fig. 4.
Optimized configuration of ivermectin with four residues of S protein which favor for hydrogen bonding with ivermectin. The hydrogen bonds are shown as dashed lines with their bond distances
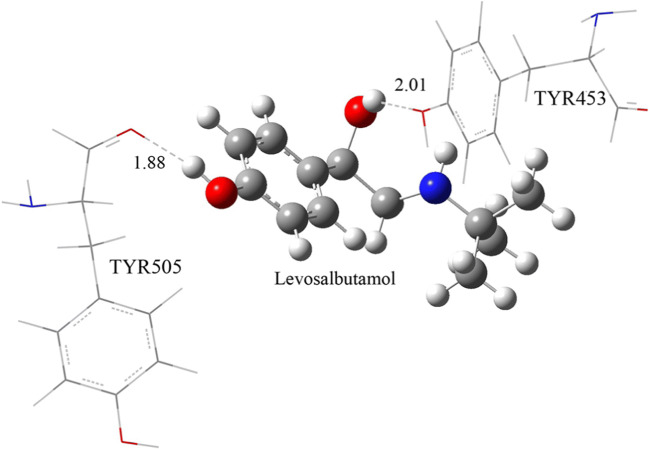
Similarly, two residues TYR453 and TYR505 with levosalbutamol were considered for DFT calculation because they formed hydrogen bonds with levosalbutamol. Their calculated binding energy is − 21.08 kcal/mol. The hydrogen atom of –OH and the oxygen atom of –CO group of TYR453 and TYR505 form hydrogen bonds with –OH groups in levosalbutamol, respectively. Their hydrogen bond distances are 2.01 Å and 1.88 Å, respectively (Fig. 5).
Fig. 5.
Optimized configuration of levosalbutamol with two residues of spike protein which favor for hydrogen bonding with levosalbutamol. The hydrogen bonds are shown as dashed lines with their bond distances
The amount of charge transfers between two interacting species that occurs for electron transfer establishes the strength of the interaction between them. We have analyzed NBO charge transfer to compute the number of charges transferred by ivermectin and levosalbutamol from or to amino acid residues. Ivermectin detracts 0.01123e− from four residues (LEU492, GLN493, GLY496, and TRY505) of S protein. On the other hand, levosalbutamol donates 1.06344e− to two residues (TYR453 and TYR505).
Stability of spike protein bound with ivermectin and levosalbutamol during MD simulation
To study the stability of the initial conformation of the docked complex in an aqueous solution, the dynamics of the complex such as RMSD (root mean square deviation) and RMSFs (root mean square fluctuations) were examined. For this, a 100-ns-long simulation was performed on the docked structure of the spike protein-ligand complexes.
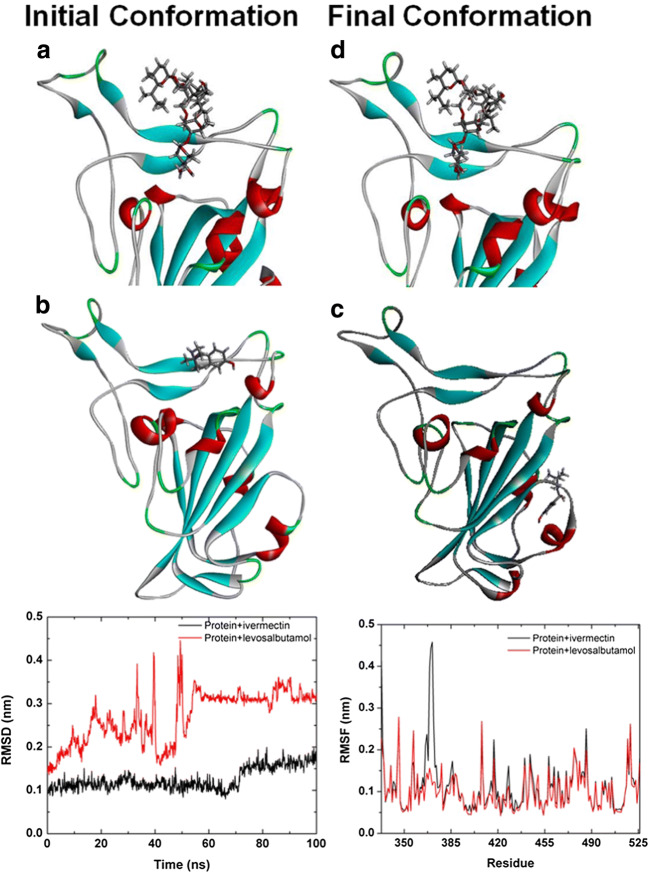
The RMSD computed for the backbone of protein (Fig. 6) shows the relative stability of the spike protein bound with ivermectin and levosalbutamol. The RMSD values for all structures were calculated using the mass-weighted least-squares fitting method. In Fig. 6, the black color represents the RMSD of the backbone of the protein bound with ivermectin, which reaches equilibrium within 2.0 ns and fluctuates between 0.9 to 0.12 nm up to 70 ns. During this period, the average RMSD value is ~ 0.11 nm. After 70 ns, the protein depicts a sudden drift towards a little higher deviation at ~ 0.18 nm. During this period, the RMSD fluctuates from 0.12 to 0.17 nm and the average RMSD is 0.15 nm. This lower deviation indicates the stability of protein bound with ivermectin in the active site throughout the simulation. On the other hand, the RMSD (red color) shows a large fluctuation for the protein bound with levosalbutamol. This is because levosalbutamol moves away from the active site of the spike protein. Therefore, it confirms that ivermectin can bind strongly in the active site of spike protein than levosalbutamol.
Fig. 6.
Initial and final snapshots of spike protein bound with ivermectin (a and d) and levosalbutamol (b and c). Bottom left shows the RMSD of protein backbone bound with ivermectine (black) and levosalbutamol (red). Bottom right figure shows the RMSF of protein bound with ivermectin (black) and levosalbutamol (red)
To understand the dynamics in more details, we compute the time-averaged RMSF (Fig. 6) for the spike protein bound with ivermectin (black color) and levosalbutamol (red color) at 300 K. The RMSF is used to evaluate the flexibility of the residues of protein which reveals the extent to which fluctuations vary along the protein chain before and after binding with ivermectin. The RMSF values of LEU492, and GLY496 are found close to the baseline (~ 0.06 nm) except GLN493 and TRY505 (< 0.15 nm). This small fluctuation of these four important amino acids will be beneficial for binding ivermectin in the S protein’s active site. The large fluctuation (~ 0.45 nm) is found for amino acids 369–371 which are relatively far from the active site of ivermectin. In the case of spike protein bound with levosalbutamol, the RMSF values for TYR453 and TYR505 are found 0.06 and 0.1, respectively.
Conclusion
Using three different computational modeling techniques, we have investigated the binding mechanism of two FDA-approved drugs (ivermectin and levosalbutamol) in the active site of the spike protein of SARs-CoV-2 where hACE2 binds. Molecular docking results predict that ivermectin shows a large binding affinity for spike protein (− 9.0 kcal/mol) compared to levosalbutamol (− 4.1 kcal/mol). Ivermectin binds with LEU492, GLN493, GLY496, and TRY505 residues in spike protein which favor hACE2 to bind in the active site. Ivermectin binds with these residues through hydrogen bonds. Levosalbutamol binds with TYR453 and TYR505 residues. Using density functional theory (DFT) studies, we have calculated the binding energy between ivermectin and four residues in the active site − 22.4 kcal/mol. In the case of levosalbutamol with two residues, it is − 21.08 kcal/mol. The natural bond orbital (NBO) charge analysis has been performed to estimate the amount of charge transfer that occurred by two drugs during interaction with residues. Ivermectin detracts 0.01123e− from four residues (LEU492, GLN493, GLY496, and TRY505) of the spike protein. On the other hand, levosalbutamol donates 1.06344e− to two residues (TYR453 and TYR505). Molecular dynamics (MD) study confirms the spike protein’s stability with ivermectin through RMSD and RMSF analyses. During a 100-ns-long MD simulation, ivermectin was bound in the spike protein’s active site. On the other hand, levosalbutamol moves away from that site. Three different computer modeling techniques confirm that ivermectin is more stable than levosalbutamol in the active site of spike protein where hACE2 binds. Therefore, ivermectin can be a suitable inhibitor for SARS-CoV-2 to enter into the human cell through hACE2.
Author contributions
Md. Jahir Raihan performed data analysis and figure preparation. Study conception, design, and manuscript writing were performed by Joyanta K. Saha.
Funding
This study was supported by Research Grant (No. 16-008 RG/CHE/AS_I – FR3240293317) funded by The World Academy of Sciences (TWAS). The authors thank TWAS for this generous support.
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J. V. Coronaviridae Study Group of the International Committee on Taxonomy of, The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang P, Wang X. COVID-19: a new challenge for human beings. Cell Mol Immunol. 2020;17:555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagar M, Ahmed HA, Aljohani G, Alhaddad OA (2020) Investigation of some antiviral N-heterocycles as COVID 19 drug: molecular docking and DFT calculations. Int J Mol Sci 21 [DOI] [PMC free article] [PubMed]
- 4.Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi C, Sun X, Ye J, Ding L, Liu M, Yang Z, Lu X, Zhang Y, Ma L, Gu W, Qu A, Xu J, Shi Z, Ling Z, Sun B. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol. 2020;17:621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ragia G, Manolopoulos VG (2020) Inhibition of SARS-CoV-2 entry through the ACE2/TMPRSS2 pathway: a promising approach for uncovering early COVID-19 drug therapies. Eur J Clin Pharmacol [DOI] [PMC free article] [PubMed]
- 7.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S.S. Ahmad, M. Sinha, K. Ahmad, M. Khalid, I. Choi (2020) Study of Caspase 8 Inhibition for the Management of Alzheimer’s Disease: A Molecular Docking and Dynamics Simulation, Molecules, 25:2071. [DOI] [PMC free article] [PubMed]
- 9.Touret F, Gilles M, Barral K, Nougairède A, van Helden J, Decroly E, de Lamballerie X, Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep. 2020;10:13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jat KR, Khairwa A. Levalbuterol versus albuterol for acute asthma: a systematic review and meta-analysis. Pulm Pharmacol Ther. 2013;26:239–248. doi: 10.1016/j.pupt.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. doi: 10.1063/1.464913. [DOI] [Google Scholar]
- 12.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- 13.Saha JK, Hossain MS, Ghosh MK. DFT study of response mechanism and selectivity of poly(3,4-ethylenedioxythiophene) towards CO2 and SO2 as gas sensor. Struct Chem. 2019;30:1427–1436. doi: 10.1007/s11224-018-1272-4. [DOI] [Google Scholar]
- 14.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams FD, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery Jr JA, Peralta JE, Ogliaro F, Bearpark MJ, Heyd JJ, Brothers EN, Kudin KN, Staroverov VN, Keith TA, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, Fox DJ (2016) Gaussian 16, Wallingford, CT
- 15.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 16.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 17.Yuan X-H, Wang Y-C, Jin W-J, Zhao B-B, Chen C-F, Yang J, Wang J-F, Guo Y-Y, Liu J-J, Zhang D, Gong L-L, He Y-W. Structure-based high-throughput epitope analysis of hexon proteins in B and C species human adenoviruses (HAdVs) PLoS One. 2012;7:e32938. doi: 10.1371/journal.pone.0032938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.