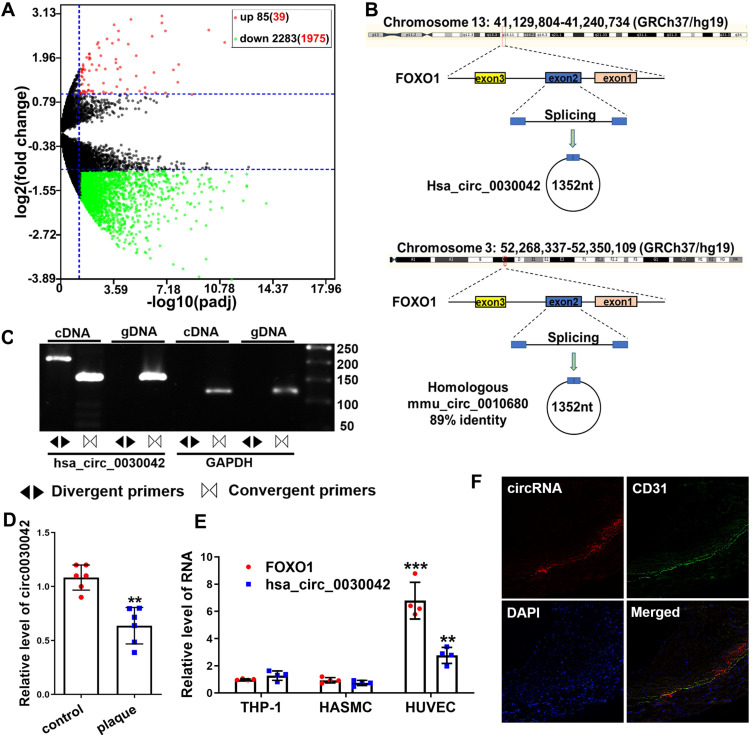
Figure 1.
circRNA sequencing uncovered CHD-related hsa_circ_0030042, which was validated using qRT-PCR in human atherosclerotic plaques. A. Volcano plot of sequenced PBMC circRNAs. X-axis: padj expressed as -log10; Y-axis: Fold change expressed as log2. The vertical line represents a padj of 0.05. The number in brackets indicated circRNAs having annotations in circRNAs. Compared with the control, the red points represent circRNAs with a fold change ≥ 2 and padj<0.05, and the green points represent circRNAs with a fold change ≤ 0.5 and padj<0.05. The horizontal line represents a padj of 0.05. Data were performed in DEseq2 R package and the padj were the p-value adjusted using the Benjamini-Hochberg procedure to control the false discovery rate (FDR). B. An illustration of the production of hsa_circ_0030042 and its homologous circRNA mmu_circ_0010680 in the mouse. C. Specific divergent primers amplified hsa_circ_0030042 in cDNA but not genomic DNA (gDNA) in HUVECs. GAPDH was used as a linear control. D. qRT-PCR validated hsa_circ_0030042 in human coronary arteries with plaques or matched controls without plaques. Data are presented as mean ± SD. Student's t-test. **p < 0.01. n = 6 pairs. E. FOXO1 mRNA and hsa_circ_0030042 levels in different vascular cells. Data are presented as mean ± SD. Student's t-test. Compared with THP-1, **p < 0.01, ***p < 0.001. n = 4. F. The location of hsa_circ_0030042 and endothelial cells were co-detected with FISH.