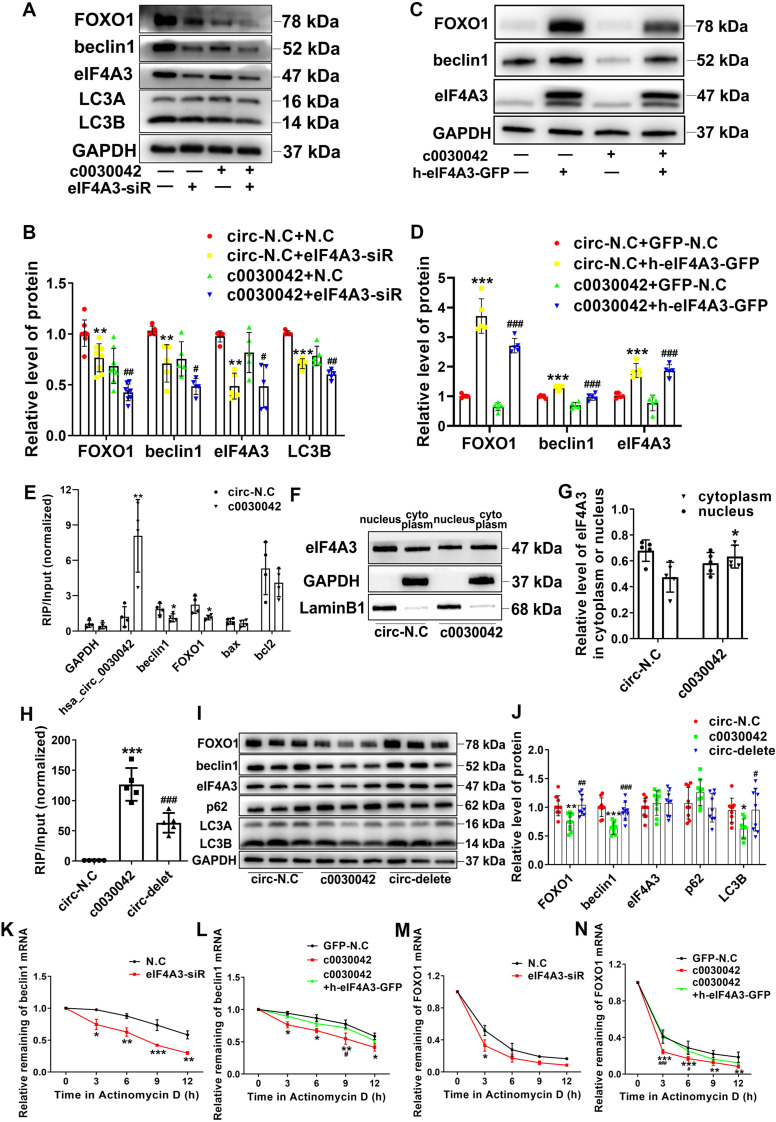
Figure 4.
hsa_circ_0030042 inhibits excessive autophagy through obstructing the recruitment of eIF4A3 to beclin1 and FOXO1 mRNA. A & B. Stable hsa_circ_0030042 overexpression HUVECs (c0030042) and empty vector transfected HUVECs (circ-N.C) were transiently transfected with negative control (N.C) and eIF4A3 siRNA for 48 h. FOXO1, beclin1, eIF4A3, and LC3B protein were quantified using western blot. One-way ANOVA, compared with circ-N.C+N.C, **p < 0.01, ***p < 0.001; compared with c0030042+N.C, #p < 0.05, ##p < 0.01. n ≥ 5. C & D. eIF4A3 lentivirus (h-eIF4A3-GFP) and empty GFP lentivirus (GFP-N.C) were transfected into c0030042 and circ-N.C, respectively. FOXO1, beclin1 and eIF4A3 were detected using western blot. Compared with circ-N.C+GFP-N.C, ***p < 0.001; compared with c0030042+GFP-N.C, ###p < 0.001. n = 5. E. RNA immunoprecipitation (RIP) of eIF4A3 from c0030042 and circ-N.C group. hsa_circ_0030042, beclin1, FOXO1, Bax, Bcl2, and GAPDH mRNA bound to eIF4A3 were determined using qRT-PCR. Input of c0030042 was normalised to that of the circ-N.C group. Student's t-test. *p < 0.01, **p < 0.01. n = 4. F & G. Western blot showing relative level of eIF4A3 in the cytoplasm or nucleus in the c0030042 and circ-N.C groups (F), and the ratio was quantified (G). Student's t-test. *p < 0.05. n = 5. H. RIP of eIF4A3 from circ-N.C, c0030042, and circ-delete groups. The eIF4A3-bound hsa_circ_0030042 hsa_circ_0030042 was determined using qRT-PCR. Input of c0030042 and circ-delete was normalised to that of the circ-N.C group. One-way ANOVA, compared with circ-N.C, ***p < 0.001; compared with c0030042, ###p < 0.001. n = 5. I & J. Western blot showing the relative level of FOXO1, beclin1, eIF4A3, p62 and LC3B. One-way ANOVA. Compared with circ-N.C, *p < 0.05, **p < 0.01, ***p < 0.001; compared with c0030042, #p < 0.05, ##p < 0.01, ###p < 0.001. n = 9. K - N. After Actinomycin D treatment, the mRNA stability of beclin1 and FOXO1 in indicated cells was determined using qRT-PCR. Two-way ANOVA. Compared with N.C, *p < 0.05, **p < 0.01, ***p < 0.001; compared with c0030042+h-eIF4A3-GFP, #p < 0.05, ###p < 0.001. n = 4. Data are presented as mean ± SD.