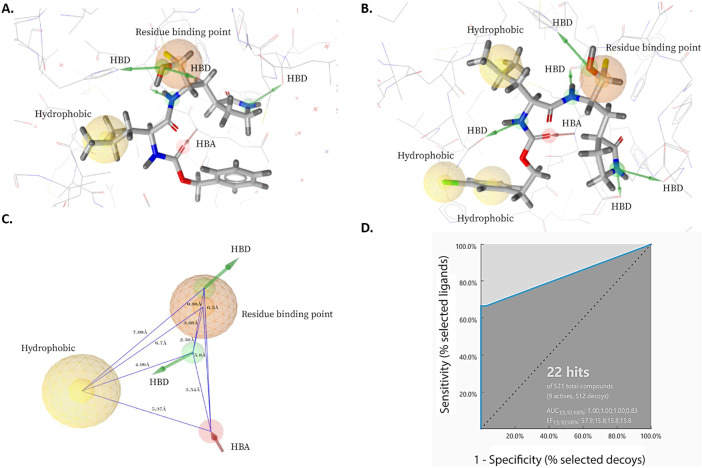
Fig. 3.
Structure-based pharmacophore models derived from X-ray structures of MERS-CoV 3CLpro enzyme in complex with (A) GC813 (PDB: 5WKK) and (B) GC376 (5WKJ). (C) A model of shared features pharmacophore of A and B. The pharmacophore features were represented in LigandScout by color codes in which, residue binding point, hydrophobic, hydrogen bind donor and hydrogen bond acceptor are depicted as orange sphere, yellow sphere, green arrow and red arrow, respectively. HBD and HBA stand for hydrogen bond donor and hydrogen bond acceptor, respectively. (D) Receiver operating characteristic (ROC) curve validation of the 3D structure-based pharmacophore model.