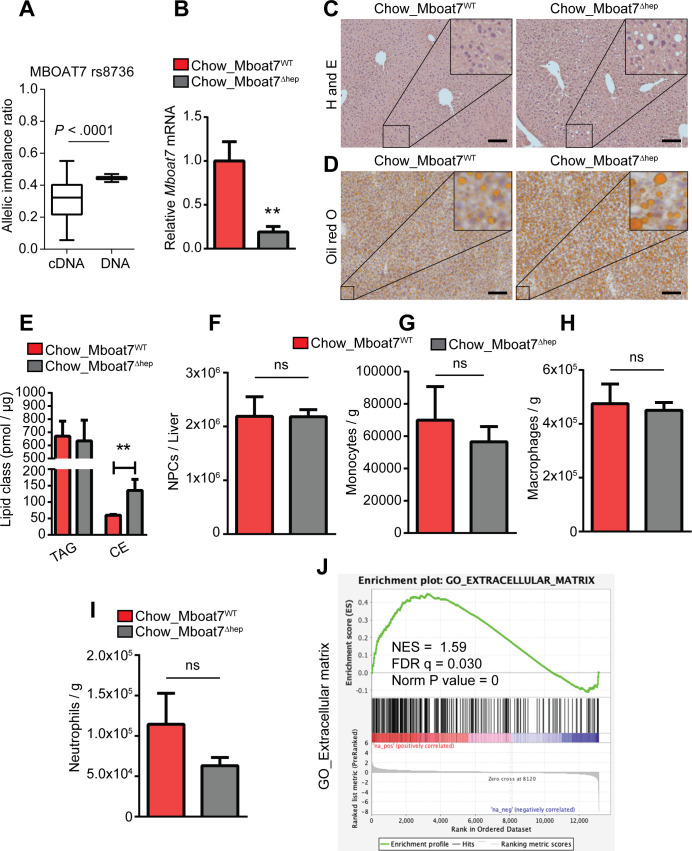
Figure 1.
Allelic imbalance and spontaneous steatosis phenotype of Mboat7Δhep in 10 weeks old mice fed with normal diet. (A) Relative allelic hepatic mRNA expression of MBOAT7 transcripts in rs641738C>T heterozygous individuals using the cSNP rs8736 that is in LD to rs641738T. Shown is the ratio of mutation carrying transcripts compared with transcripts bearing the ancient allele (left column). DNA (right column) was used as a control. n=37. (B–J) Data derived from Mboat7WT and Mboat7Δhep mice which were fed a normal diet. Data are presented as mean±SEM. **P<0.01 (Mann-Whitney U test). (B) mRNA expression of Mboat7 was determined by qRT-PCR in Mboat7WT (n=6) and Mboat7Δhep (n=7) mice. (C) H&E staining of livers from 10-week-old Mboat7WT and Mboat7Δhep mice. (D) Oil red O staining of livers from 10-week-old Mboat7WT and Mboat7Δhep mice. Scale bars, 100 µm. (E) Hepatic-concentrations of triglyceride (TAG) and cholesterol ester (CE) (n=5 mice). (F–I) Flow cytometry analysis was performed in non-parenchymal cells (NPCs). (F) Total number of NPCs (n=6–7 mice). (G–I) Hepatic number of monocytes (CD45+/Ly6G-/CD11b+/F4/80-), macrophages (CD45+/Ly6G-/CD11b+/F4/80+) and neutrophils (CD45+/Ly6G+/CD11b+) in livers of Mboat7WT and Mboat7Δhep mice (n=6–7 mice). (J) liver RNA sequencing in Mboat7WT and Mboat7Δhep mice. GSEA shows a positive correlation between ECM related genes and Mboat7 deficiency. qRT-PCR, quantitative real time-polymerase chain reaction; CE, cholesteryl ester; ECM, extracellular matrix; GSEA, gene set enrichment analysis; LD, linkage disequilibrium; MBOAT7, membrane-bound O-acyltransferase domain containing 7; NS, not significant; SEM, SE of the mean.