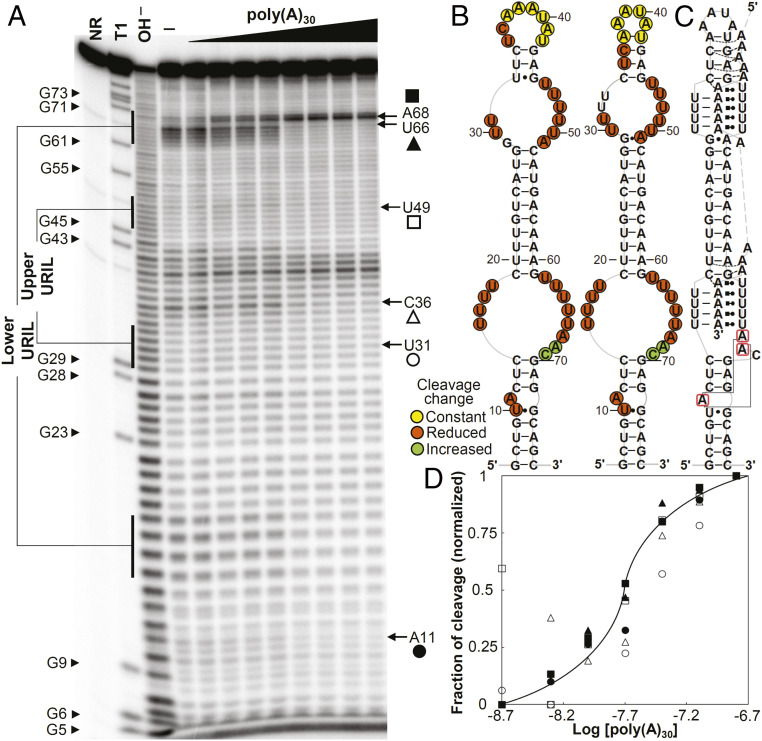
Fig. 6.
In-line probing analyses of the wild-type dENE. (A) In-line probing analyses of 32P 5ʹ end-labeled dENE in the absence (—) and presence of varying concentrations of poly(A) (2 to 320 nM) at room temperature (22 °C). Lane NR shows no reaction, lane T1 indicates RNase T1-digested RNA (G-specific cleavage), and lane OH− shows alkaline-mediated partial digestion. (B) Sequence and secondary structure models for the wild-type dENE (derived from in-line probing data in the absence of poly(A) on the Left and the predicted from comparative sequence analysis (11) on the Right). Nucleotide linkages that undergo constant, reduced, or increased cleavage upon poly(A) binding as determined by their relative band intensities in A are circled in yellow, red, or green, respectively. (C) Schematic of the poly(A)-bound dENE structure derived from the crystal structure (14). Long-range interactions between the A-triad nucleotides (boxed in red) are indicated by solid gray lines. (D) Dependence of the fraction of cleavage at selected sites (marked in A) in the lower dENE domain (closed symbols) and the upper dENE domain (open symbols) on poly(A) concentration. Calculation of fraction modulated is described in Materials and Methods. Data from selected cleavage sites were plotted against the logarithm of poly(A) concentration showing an apparent KD of ∼15 to 30 nM. The solid line indicates a theoretical binding curve for a 1:1 dENE-poly(A) interaction with a KD of 20 nM.