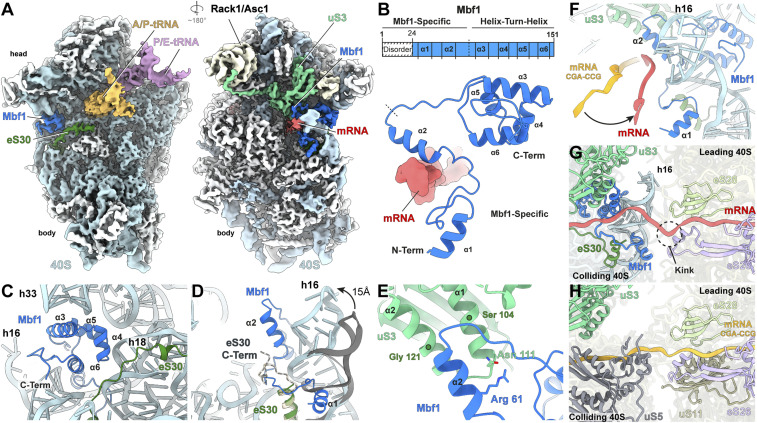
Fig. 3.
Structure of Mbf1 on the colliding ribosome. (A) Cryo-EM map of the interface (Left) and solvent (Right) view of the 40S subunit from the colliding ribosome, with 40S proteins (white), 18S rRNA (cyan), A/P-tRNA (gold), P/E-tRNA (purple), eS30 (dark green), uS3 (light green), mRNA (red) and Mbf1 (deep blue). (B) Schematic and cartoon representation of the Mbf1 molecular model (deep blue). (C) Mbf1 helix-turn-helix motif bound to the 40S subunit. (D) Refolding of h16 and eS30 C terminus destabilization on Mbf1 binding. Comparison of h16 (cyan) and eS30 (dark green) in the Mbf1-bound colliding ribosome to h16 (gray) and eS30 (light gold) in the colliding ribosome of an Mbf1-lacking disome (Protein Data Bank [PDB] ID 6SNT) (24) (E) Close-up view of uS3 and Mbf1 helix 2 interactions. (F) Comparison of the path of the mRNA (red) in the Mbf1-bound structure compared to the mRNA (orange) in a colliding ribosome of an Mbf1-lacking disome (PDB ID 6I7O) (23). (G and H) The mRNA path between the 40S of the colliding ribosome and the 40S of the leading ribosome within the Gcn1-disome (G) and the CGA-CCG stalled disome (PDB ID 6I7O) (H) (23). Components interacting with the mRNA at the 40S-40S interface are shown for each disome, respectively.