Fig. 1.
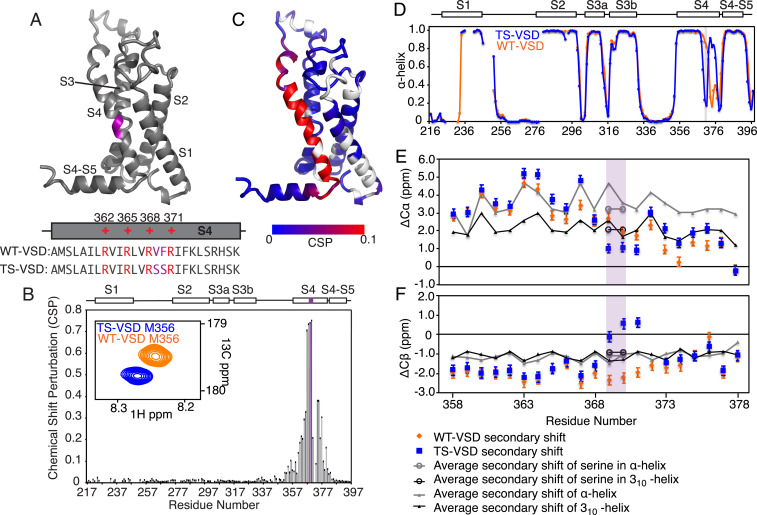
Serine mutations in S2bot alter the structure of Shaker-VSD. (A) The two-amino-acid S2bot mutation (V369F370 to S369S370) is highlighted in magenta on the homology model of isolated Shaker-VSD and in the S4 helix sequence (Lower Inset, voltage-sensing arginines in red). (B) CSP between WT-VSD and TS-VSD was calculated as described in Materials and Methods using the well-resolved 3D-HNCO spectra at 45 °C. The graph is numbered according to the full-length Shaker sequence, with the S2bot mutation (magenta) and transmembrane helices S1 to S4 indicated at the top. An overlay of the WT-VSD and TS-VSD peaks (A, Inset) corresponding to M356 in the 3D-HNCO illustrates the significance of a CSP value of 0.1. (C) The CSP data are plotted on the homology model of the isolated Shaker-VSD (blue to red color scale as shown, unassigned residues in white). Significant CSP ≥ 0.1 (red in B) is observed at the site of mutation and extending along the S4 transmembrane helix and into the adjoining loops. Similar results are observed at 28 °C (SI Appendix, Fig. S2). (D) The α-helical propensity of WT-VSD (orange) and TS-VSD (blue) was calculated using TALOS+ (25) analysis of the backbone NMR chemical shifts (HN, N, Co, C-α, and C-β). The scale extends from 0 (no helical propensity) to 1.0 (α-helix). The gaps in data reflect residues without backbone assignments. TS-VSD is generally similar to WT-VSD except for a loss of α-helical propensity at the S2bot mutation site (purple shading) and extending toward the C-terminal end of S4. Almost identical predictions are obtained with PECAN (26) (SI Appendix, Fig. S2). The raw data underlying the secondary structure analysis include the C-α (E) and C-β (F) secondary chemical shifts, which are shown for the S4 transmembrane helix of WT-VS (orange) and TS-VSD (blue) plotted against the average secondary chemical shifts for the corresponding amino acid sequence in a fully α-helix (gray) or 310-helix (black) conformation as extracted from the PACSY database (27).