Fig. 2.
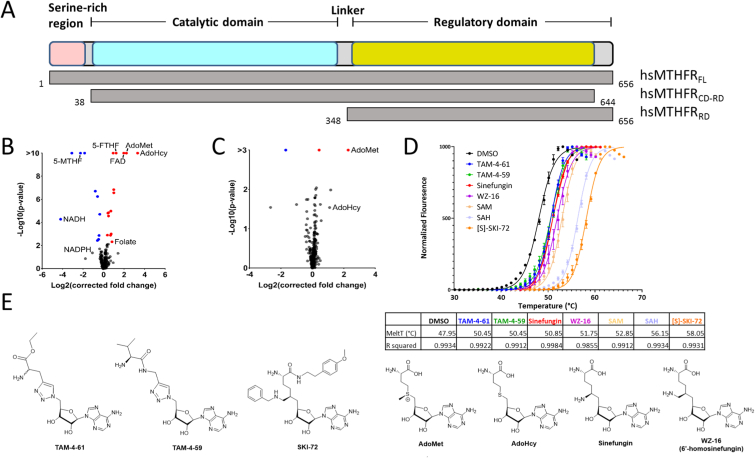
Identification of compounds bound by the regulatory domain of MTHFR. A. Schematic domain organisation of MTHFR depicted above in colour with protein constructs used in this study visualized below in grey. Amino acid boundaries for each are given, based on NP_005948. B. The hsMTHFRCD-RD–metabolite interactome as determined by MIDAS. AdoMet, AdoHcy, FAD, folate, and 5-FTHF were enriched and 5-MTHF, NADH, and NADPH were depleted. C. The hsMTHFRRD–metabolite interactome as determined by MIDAS. AdoMet and AdoHcy were enriched. B and C, red data points indicate significantly enriched metabolites and blue data points indicate significantly depleted metabolites. The cut-off for significance was p < 0.05 and q < 0.1. D. Above: Representative curves of the differential scanning fluorimetry binding assay for hsMTHFRRD in the absence (DMSO) or presence of 250 μM of each compound. Each curve represents n = 2, replicates pooled and fitted. Below: Table indicating the average melting point and goodness of fit for each curve. E. Structures of the compounds indicated in panel D.