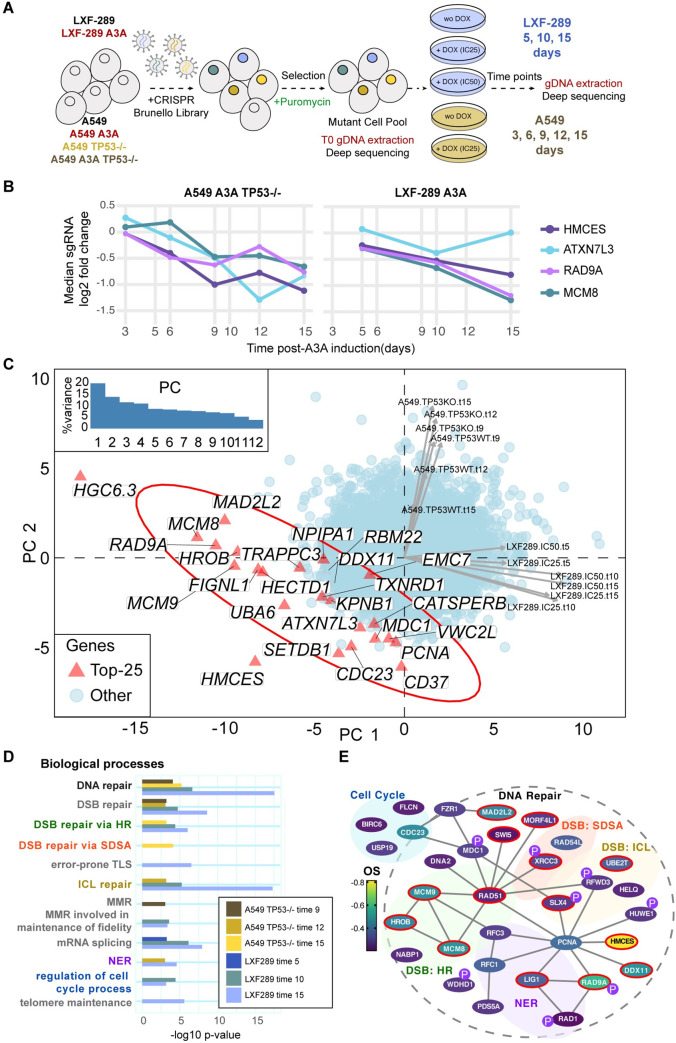
Fig 2. CRISPR/Cas-9 genetic screen indicates HMCES and other DNA repair genes as vulnerabilities of A3A-expressing cells.
(A) Experimental design using the Brunello genome-wide library [50]. (B) Depletion of the sgRNAs targeting top 4 genes upon A3A overexpression, as prioritized by the overall A3A conditional essentiality score: LFC across 3 time points of the LXF-289 cell line and the 3 latest time points of the A549TP53−/− cell line. The y-axis shows the median of the 4 sgRNAs per gene. (C) PC analysis of A3A conditional LFC scores for all genes across all 12 experimental conditions (see labels next to arrows, which show loadings of the conditions on PC1 and PC2; “KO” implies TP53−/− and “WT” TP53 wild-type A549 cell line; numbers in labels are the time points; “IC25” and “IC50” are 2 concentrations of DOX in the LXF-289 cell line). The top 25 genes, prioritized by the same A3A conditional score as in panel B, are highlighted on the figure. Inlay shows a scree plot, with the amount of variance explained by the 12 PCs. The LXF-289-specific HGC6.3 hit is likely an artefact (S1 Table; see Results text). LFC for all genes/sgRNAs are included in S2 Table and S2 Data. LFC for sgRNAs (from the top 100 genes plus nontargeting), shown in S2A–S2C Fig, are included in S3 Data. (D) Gene Ontology enrichment analysis of the top hits in the 6 experiments considered for the overall A3A conditional score (as in panel B). Plot shows −log10 p-value (unadjusted) from the GORILLA server. Underlying data and full Gene Ontology analysis and category names are provided in S3 Table. (E) Network schematic of cell cycle and DNA repair–related genes from the top 300 hits in the screen (OS). Genes identified in the Gene Ontology analysis and appearing in the top 300 genes by OS are shown. Color denotes the OS, lines indicate physical interactions (thebiogrid.org) [52], and a red border indicates they were identified in the Gene Ontology analysis in both cell lines. Those gene products identified as PIKK (ATM, ATR, and DNA-PKcs) targets are indicated with a purple “P,” data from PhosphoSitePlus [53]. Numerical data for all graphs in the figure are provided in S3 Table and S2 Data. A3A, APOBEC3A; DSB, double-strand break; HR, homologous recombination; ICL, interstrand crosslink; LFC, log2 fold change; MMR, mismatch repair; NER, nucleotide excision repair; OS, overall score; PC, principal component; PIKK, PI-3 kinase-like kinase; SDSA, synthesis-dependent strand annealing; sgRNA, single gRNA; TLS, translesion synthesis.