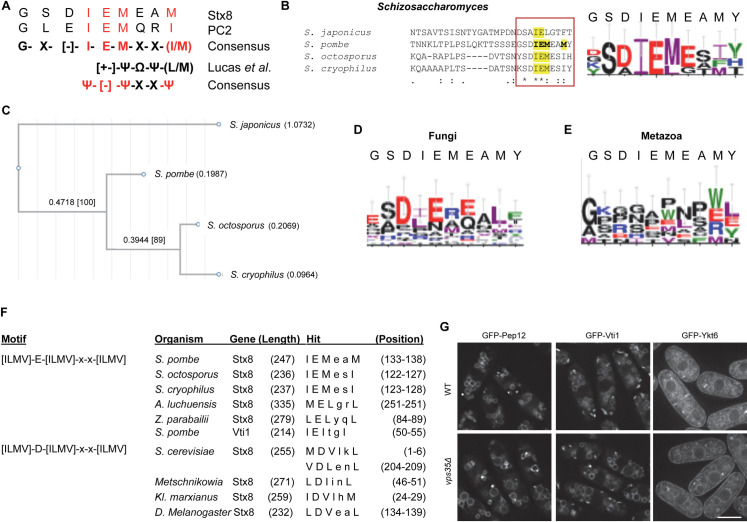
Fig 8. Conservation of the Stx8 IEMeaM motif.
(A) Comparison between the motifs in Schizosaccharomyces pombe Stx8 and human PC2, and the consensus motif defined by Lucas et al [26] where Ψ was hydrophobic residue with large aliphatic chain; [+-] a charged residue; and Ω a bulky hydrophobic (aromatic) residue [26]. In the case of Stx8 and PC2, where the charged residue is an E, the consensus at this position is [–]. The residues essential for function are in red font. (B) Left panel: BLASTP alignment of Fsv1/Stx8 proteins from the Schizosaccharomyces species. The square denotes the region where the IEMeaM motif is located. Right panel: Weblogo showing the conservation of the residues denoted by the square in the left panel. The S. pombe sequence is indicated at the top. E, D in red; H, K, and R in blue; F, Y, and W in green; and I, L, M, and V in purple. (C) Phylogram of the Fsv1/Stx8 sequences aligned with clustal W (B). (D) The same details as in (B), but the analysis was extended to 19 fungal Stx8 sequences. (E) The same details as in (B), but the Weblogo was deduced from the S. pombe, Homo sapiens, Pan troglodytes, Rattus norvegicus, Mus musculus, Danio rerio, Drosophila Melanogaster, and Arabidopsis thaliana Syntaxin 8 sequences. (F) ScanProsite hits detected by scanning for the indicated motifs. (G) Localization of the indicated SNAREs in WT and vps35Δ.