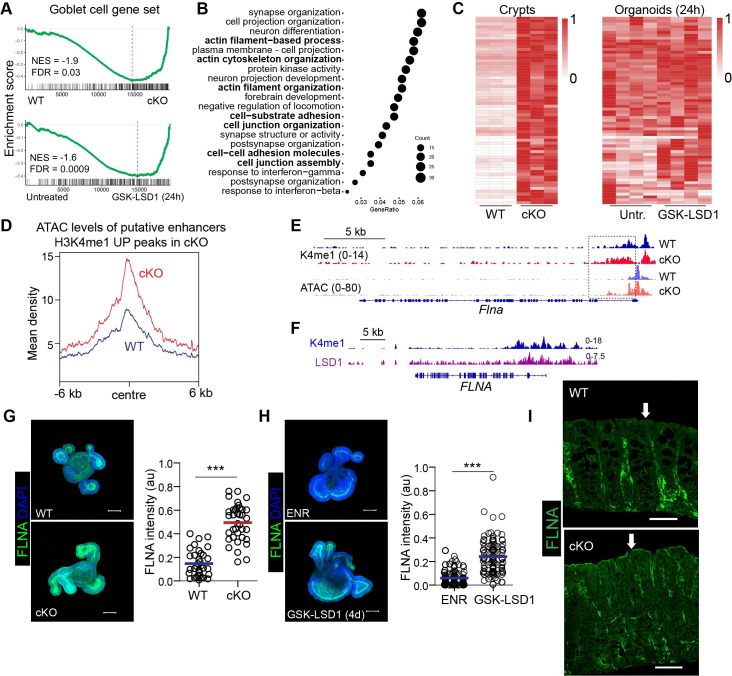
Fig 6. LSD1 epigenetically controls modulators of the cytoskeleton.
(A) Application of GSEA of a goblet cell gene set in WT and cKO small intestine RNA seq, and untreated vs GSK-LSD1 (24h) treated organoids RNA-seq data. (B) Top 500 genes upregulated in cKO crypts were selected and unbiased association with the top 20 GO terms is displayed. (C) Genes taken from in 6B bold-selected GO terms are displayed in a heatmap using Transcripts Per Million (TPM) values scaled to the max of the gene comparing WT and cKO crypts (left) and untreated and GSK-LSD1 (24h) treated organoids (right). (D) Mean ATAC signal of normalized peak density surrounding previously identified H3K4me1 peaks that are significantly up in cKO crypts compared to WT crypts. Representative graph from n = 2. (E) Representative Integrative Genomics Viewer (IGV) tracks of H3K4me1 and ATAC signal at the Flna locus (n = 2). (F) IGV tracks of H3K4me1 and LSD1 levels at the FLNA locus in a human cancer cell line. (G) Representative confocal images of FILAMIN A (FLNA) stained in WT and cKO organoids. Quantification of FLNA intensity in WT and cKO organoids. (H) WT organoids were untreated (ENR) or treated (GSK-LSD1- 5 μM) for 4 days and then stained for FLNA (green). DAPI was used as a counter stain (blue). Scale bar = 50 μm Organoids were quantified for FLNA fluorescent intensity. (I) FLNA staining in paraffin embedded colon sections of naive WT and cKO mice. White Arrow indicates the increased FLNA expression on the apical side of colonic crypts. Scale bar = 50 μm. Unpaired Student’s t test was performed to observe significant differences. *** P ≤ 0.001.