Summary:
Eukaryotic cells integrate multiple quality control (QC) responses during protein synthesis in the cytoplasm. These QC responses are signaled by slow or stalled elongating ribosomes. Depending on the nature of the delay, the signal may lead to translational repression, messenger RNA decay, ribosome rescue, and/or nascent protein degradation. Here, we discuss how multiple pathways that converge on mRNA decay rely on sensing the ribosome in a troubled or aberrant state and how the structure and composition of this aberrant complex determines the downstream quality control pathways that ensue.
Introduction:
The proteome depends on the robust and precise translation of mRNAs. As such, cells maintain accurate translation of “good” mRNAs and minimize output from “problematic” mRNAs. Translation initiates through scanning of the small (40S) subunit of the ribosome accompanied by multiple initiation factors along the 5’ leader of the mRNA until the AUG start codon is recognized. Then the large (60S) subunit is recruited and translation begins. The 80S ribosome then moves along the open reading frame three nucleotides at a time, deciphering the codons and forming peptide bonds. Finally, the ribosome recognizes a stop codon (UAG, UGA or UAA) and terminates translation. The elongation rate of the ribosome depends on multiple factors including the abundance of the charged tRNA in the cell, the ability of the incoming charged-tRNA to engage the respective codon, the ability of the ribosome to synthesize the peptide bond, and the ability of the ribosome to translocate along the mRNA and to advance the nascent polypeptide through the peptide exit tunnel. Disruption of any of these steps can impact the state of the ribosome and signal QC pathways to mitigate the effects of elongation distress. As we will discuss throughout this review, the state of a distressed elongating ribosome can be characterized in many ways including rotation status, tRNA occupancy, proximity to the end of the mRNA, and collision status. Importantly, the molecular specifics of the elongation disruption determine the precise pathways that are signaled.
Messenger RNA (mRNA) serves as a molecular middleman between the hereditary information stored in DNA and the cellular activity carried out by proteins. In eukaryotes, RNA polymerase II transcribes DNA into precursor mRNAs, which are processed into mature mRNA containing a 5’ cap, a 5’ leader sequence, an open reading frame (ORF), a 3’ untranslated region (UTR), and a poly-adenosine tail (poly-A). The ORF of the mRNA is then translated by the ribosome into protein. In dividing cells, mRNAs are generally decayed by what we refer to as normal or canonical pathways, while problematic RNAs are rapidly decayed by separate mechanisms. Numerous studies over the past decade suggest that generally inefficient translation (characterized by slow moving ribosomes) signals normal mRNA turnover, whereas wholly unproductive translation (characterized by stalled ribosomes) signals more dramatic outcomes including translational repression, ribosome rescue, nascent peptide degradation and mRNA decay (Hanson and Coller, 2017; Inada, 2020).
Quality control (QC) responses are often triggered as a result of ribosomes encountering sequences or environmental insults that lead to a slowing in translation. Examples of sequence-induced problems include extremely poor codon content, errors in transcription, or errors in pre-mRNA processing including splicing and polyadenylation, whereas environmental damage could include exposure of cells to oxidizing agents or UV irradiation, both of which damage nucleotides (Gordon et al., 2015; Yan and Zaher, 2019). An interesting set of observations to consider is that slow ribosomes simply signal canonical mRNA decay whereas errors such as premature polyadenylation within the ORF signal more comprehensive QC. The cell clearly distinguishes between different types of translation elongation difficulties and leverages an appropriate response. Slowly decoded codons do not signal a crisis meriting nascent peptide degradation; logically the cell needs the protein output of the ORF. Instead, slow codons are taken advantage of by the cell for the regulation of mRNA turnover (Hoekema et al., 1987; Caponigro, Muhlrad and Parker, 1993; Presnyak et al., 2015). By contrast, when there are more substantive problems with the mRNA such that the ORF is unlikely to generate a full length protein product, then the cell targets the incomplete nascent peptide for decay and often inhibits further rounds of initiation before degrading the mRNA to minimize protein output (Brandman and Hegde, 2016). We know these steps are critical since deletion of factors involved in these processes lead to the accumulation of protein aggregates and cell death in yeast (Bengtson and Joazeiro, 2010) and neurodegeneration in mammals (Ishimura et al., 2014; Martin et al., 2020).
In this review, we provide an overview of recent work in the field exploring both normal mRNA turnover and problematic mRNA quality control. Normal mRNA turnover depends on multiple pathways that are critical for overall protein expression; similarly, mRNA quality control depends on multiple overlapping pathways that process trapped ribosome complexes and target incomplete toxic proteins and mRNAs for decay. We discuss how these two major processes of mRNA turnover and mRNA quality control initiate at the ribosome and distinguish at a molecular level between normal (possibly slowly) elongating ribosomes and terminally stalled ribosomes to trigger a measured response.
Canonical mRNA decay and connections to translation elongation
Mechanism of mRNA decay
Messenger RNA half-lives vary from seconds to >30 minutes in yeast (Chan et al., 2018) and seconds to days in mammalian cells (Tani et al., 2012) indicating that mRNA turnover plays a major regulatory role in determining the protein content of cells. The players in eukaryotic mRNA turnover have been extensively studied and are highly conserved from yeast to mammals. The majority of mRNAs are initially decayed by a process that begins with shortening of the poly-A tail (deadenylation) by both the Pan2-3 complex and the CCR4-NOT complex. These deadenylase complexes contain catalytic exonucleases at their core – Pan2 functions as the exonuclease in the Pan2-3 complex and Caf1 and Ccr4 in the CCR4-NOT complex. The predominant model in the field is that Pan2-3 trims longer poly-A tails (>150 nt in mammalian cells), while CCR4-NOT does the final trimming that leads to decapping and mRNA decay (Chen and Shyu, 2011; Wahle and Winkler, 2013; Webster et al., 2018; Yi et al., 2018). Following poly-A shortening, decapping is signaled through elaborate and likely redundant pathways involving a multitude of factors (see review (Parker, 2012)). In one possible pathway, the CCR4-NOT complex stimulates the decapping regulator Dhh1 (DDX6 in mammalian cells), a member of the DEAD Box protein family. While the mechanism is not fully established, the Not1 protein of the CCR4-NOT complex contains a mIF4G domain that, along with RNA, stimulates the ATPase activity of Dhh1 (Mugler et al., 2016). Once activated, Dhh1 interacts with or recruits Pat1 and Edc3, accessory decapping regulators, and Dcp2, the catalytic subunit of the decapping complex Dcp1/2. Dcp1/2 removes the 5’ cap allowing the major 5’ to 3’ exonuclease Xrn1 to rapidly decay the mRNA. This canonical mRNA decay occurs as a co-translational process culminating in Xrn1 closely trailing the final (5’ most) ribosome on the message (Hu et al., 2009; Pelechano, Wei and Steinmetz, 2015; Tesina, Heckel, et al., 2019). At the same time, once an mRNA is completely deadenylated in the cytoplasm, the SKI complex, consisting of a 3’ to 5’ helicase Ski2 and accessory factors Ski3, Ski7, and Ski8, binds the free 3’ end and recruits the major 3’ to 5’ decay machinery, the exosome, to degrade the message (Anderson and Parker, 1998). These two exonucleolytic machines are the major modes of cytoplasmic decay in eukaryotes and while in yeast Xrn1-mediated decay is the predominant pathway (Hu et al., 2009), the relative roles of these pathways is less certain in mammals.
Codon optimality impacts mRNA turnover
Although the major players in mRNA decay have been known for over a decade, the upstream signals for the initial deadenylation step are less well defined. An important recent discovery is that codon optimality correlates with overall mRNA decay promoted by the decapping machinery and Xrn1 (Presnyak et al., 2015). Broadly speaking, codon optimality is defined by the tRNA Adaptive Index (tAI) which is a metric that takes into account the abundance of the aminoacylated-tRNA and the strength of the tRNA anticodon to decode its cognate or near-cognate codon pair within the ribosome (dos Reis, Savva and Wernisch, 2004). While there are other related metrics that have been developed, all broadly correlate with one another and are similarly correlated with mRNA half-life in yeast (Presnyak et al., 2015). An essential role for codon optimality within a given mRNA is to allow proper folding of nascent peptide domains (Rodnina, 2016; Liu, 2020), although this will not be a focus of this review. Here, we will instead focus on how codon optimality impacts mRNA half life.
In a key publication, a genome-wide study of mRNA half lives discovered that the more optimal the codons within an mRNA, the longer the half life (Presnyak et al., 2015). This correlation that was originally observed in yeast has since been corroborated in a variety of organisms including mammals (Wu et al., 2019; Forrest et al., 2020), zebrafish (Mishima and Tomari, 2016), and even bacteria (Boël et al., 2016). Though the contributions of optimality are generally more modest in mammalian cells, genes that are more dosage-sensitive (in many cases disease relevant genes) have been shown to have a stronger selection for a specific codon content (Dhindsa et al., 2020). This dependence of mRNA half-life on codon optimality immediately implicates the ribosome as a major regulator of mRNA turnover.
Multiple forms of evidence, including in vitro translation in cell lysates, single molecule microscopy experiments, and in vivo ribosome runoff studies support the idea that ribosomes elongate more slowly along mRNAs with lower codon optimality, relative to those with optimal codons (Presnyak et al., 2015; Yu et al., 2015; Yan et al., 2016). What these data suggest then is that the rate at which a ribosome moves along an mRNA template in some way determines the rate at which the mRNA is decayed, thus providing a mechanism for cells to regulate mRNA turnover and thus steady state levels of the protein products. Reassuringly, mRNAs encoding for highly abundant, well translated proteins are typically optimally encoded and are also very stable (Presnyak et al., 2015). However, how this regulation is implemented at a molecular level has until recently been elusive.
Since codon-driven decay was initially reported, a number of canonical decay factors have been implicated as critical for sensing slow ribosomes and triggering decay. The major decapping-associated decay factor Dhh1 (DDX6 in mammals) was implicated in the process of targeting nonoptimal mRNAs for rapid decay, but the mechanism for identifying transcripts with slow translation rates was unknown (Radhakrishnan et al., 2016). Similarly, the catalytic deadenylase Caf1 (CNOT7 in mammals) was shown to specifically deadenylate nonoptimal mRNAs, but the upstream signals for this reaction were also unknown (Webster et al., 2018). Importantly, neither of these factors has been shown to bind directly to ribosomes.
Recently, the protein Not5, a member of the CCR4-NOT complex, was implicated in the decay of nonoptimal mRNAs. Moreover, through structural and biochemical studies, the N terminal domain of Not5 was shown to bind directly in the tRNA exit site, or E site, of ribosomes, in a manner incompatible with tRNA binding (Buschauer et al., 2020). This structural study also suggested that the conformation of the E site that houses the N terminal domain of Not5 is dependent on an empty A site because the E site slightly changes upon A-site tRNA incorporation (Buschauer et al., 2020). In bacteria, previous biochemical studies demonstrated that the affinity of tRNA for the ribosome in the E site is relatively low and dissociation is spontaneous (Uemura et al., 2010; Belardinelli et al., 2016). In yeast, the E-site tRNA is not thought to spontaneously release, but instead release is promoted by the elongation factor eEF3 (an ATPase) independent of aminoacyl-tRNA binding in the A site (Ranjan et al., 2020). Independent of the mechanism of E-site tRNA release, if ribosomes are slow to decode, it is more likely that the E-site tRNA will have been released, thus allowing E-site occupancy (or lack thereof) to report on slow ribosomes. As such, binding of the N terminus of Not5 in the E site is a first molecular insight into recognition of a slow moving ribosome.
What will be critical next is to understand how Not5 binding leads to the recruitment of the remainder of the decay machinery. Buschauer et al. show that in addition to Not5, the specific decay of nonoptimal mRNAs requires Not4-dependent ubiquitination of ribosomal protein S7, suggesting that the entire CCR4-NOT complex regulates decay. The CCR4-NOT complex is itself a deadenylase and interacts with a collection of decapping factors, including Dhh1, immediately providing an idea as to how decay might occur -- a plausible path includes recruitment of the CCR4-NOT complex in its entirety to the slow ribosomes through interactions with Not5 bound in the E site (see Figure 1).
Figure 1:
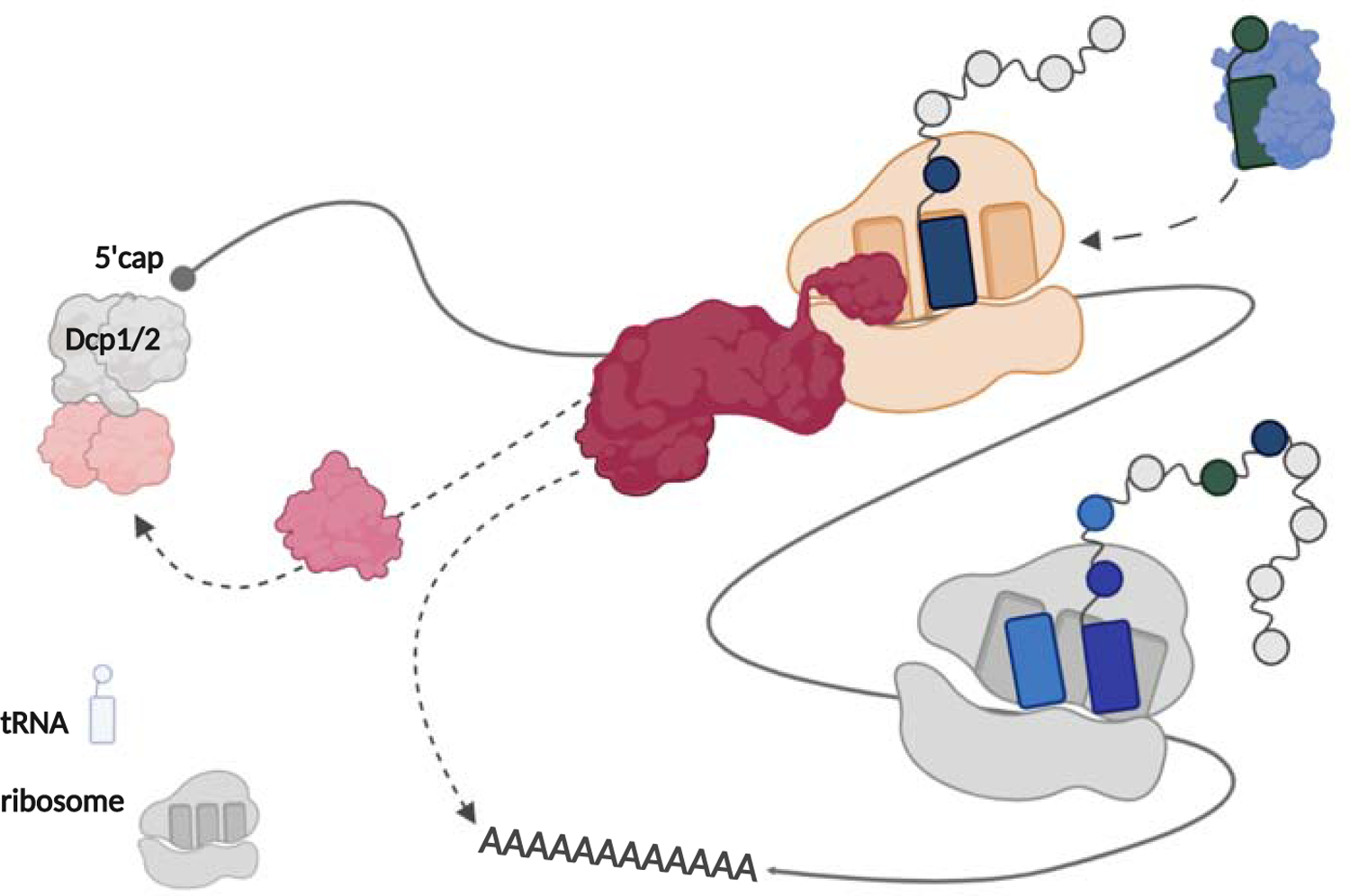
Model for mRNA decay on nonoptimal mRNA. The N-terminus of Not5 recognizes an empty E site on a slowly elongating ribosome. By recruiting CCR4-NOT, Not5 signals canonical Xrn1-mediated mRNA decay.
These studies support the idea that slowly elongating ribosomes are a key trigger for normal canonical mRNA decay and that recognition is mediated through factor binding in the E site. We know from other studies that there are multiple mechanisms for recruiting the CCR4-NOT complex to mRNAs to target them for decay, though these often involve recruitment by factors bound to the 3’ UTR of the mRNA (Webster, Stowell and Passmore, 2019). However, these latter examples do not likely involve slowly elongating ribosomes, but instead provide an example of mRNA decay that is ribosome independent. For this reason, they are not the focus of discussion here.
QC on problematic mRNAs with disruptions within the ORF
How the cell identifies mRNA with damage to the ORF:
There has been considerable focus in recent years on the cellular response to defective or damaged mRNAs. What is abundantly clear is that incomplete peptide products originating from such mRNAs are toxic to cells and that a robust quality control response has evolved in order to target such peptides for decay (Inada, 2020) – this will not be the focus of this review. Here we will focus on other QC events including mechanisms that have evolved to limit further translation on such mRNAs, to target the mRNAs for decay, and, to rescue the stuck ribosomes for re-entry into the cellular pools. The key first question is how does the cell distinguish between a normal mRNA being translated and a problematic mRNA that should be targeted by QC. To answer this, we will separately discuss two broad classes of problematic mRNA – those with ribosomes stalled within the ORF and those with ribosomes stalled in the poly-A tail.
Research over the past decade has shined light on QC events on problematic mRNAs with disruptions within the ORF. In principle, these disruptions can be encoded or environmental, though it seems likely that environmental damage is the more abundant problem faced by the cell. For example, much like DNA damage in the nucleus, RNA damage takes the form of broken, crosslinked, or chemically modified nucleotide sequences, any of which would be likely to impede elongation by the ribosome along such an mRNA template (Yan and Zaher, 2019; Yan et al., 2019; Wu et al., 2020). Encoded disruptions in the mRNA could include strong natural pauses on specific peptide motifs (Han et al., 2020; Matsuo et al., 2020), collections of rare codons (Kuroha et al., 2010; Letzring, Dean and Grayhack, 2010), or mRNA secondary structural elements (Doma and Parker, 2006), though such mRNA sequences seem to rarely occur without a corresponding regulatory purpose (Collart and Weiss, 2020).
Since damaging agents do not target a single mRNA but instead all mRNAs in the cell, and additionally can produce many off target effects, the field has largely used reporters with translationally problematic codons to mimic RNA damage. In yeast, the task is relatively simple – iterated CGA codons abruptly stall ribosomes since CGA is decoded by Arg-tRNAICG which is of unusually low abundance and possesses an ICG anticodon that decodes with a structurally perturbing I:A base pair in the wobble position (Kuroha et al., 2010; Letzring, Dean and Grayhack, 2010). In the context of these multiple challenges, decoding is slow and the iterated CGA sequence in the mRNA samples an unusual stem loop structure within the A site of the ribosome, resulting in severe steric hindrance that leads to very strong stalling of the ribosome(Tesina, Lessen, et al., 2019).
It has been more challenging in mammalian cells to identify problematic sequences that mimic RNA damage for reporter studies since in mammals the CGA codon is not decoded by an inosine-containing anticodon nor is the Arg-tRNAUCG of low abundance (Maraia and Arimbasseri, 2017). Here, poly-A sequences have been used to promote internal ribosomal stalls, though as we will discuss, these may be less good mimics of ribosomes trapped upon damaged mRNA (Arthur et al., 2015; Chandrasekaran et al., 2019). One strong stalling reporter that has been used in mammalian cells is an mRNA with a highly structured 3’ end (the MALAT sequence), though it is not clear that this 3’ terminal sequence effectively mimics an ORF-internal stalling mRNA (Hickey et al., 2020). More recently, a known natural stalling sequence, XBP1u, has been used to induce ribosomal collisions in reporter mRNA and will likely continue to be a useful tool for studying mammalian ribosomal collisions (Han et al., 2020; Sinha et al., 2020).
Using these tools, recent biochemical studies have discovered an important clue as to what distinguishes substantive ribosome stalls from simply slowed ribosomes: when ribosomes stall within an open reading frame, the upstream ribosomes collide into the stalled ribosome (Simms, Yan and Zaher, 2017), producing a novel interface that triggers the entire cascade of QC events (Juszkiewicz et al., 2018; Ikeuchi et al., 2019). We have learned a great deal about these collisions from emerging structural studies that have defined the molecular details of this interface. Structural studies reveal molecular details of the lead ribosome in an “unrotated state” with an empty A site and a peptidyl tRNA in the P site, and the lagging “colliding” ribosome in a “rotated” state, with a peptidyl-tRNA in the A-P state and an uncharged tRNA in the P-E state (Juszkiewicz et al., 2018; Ikeuchi et al., 2019). The collision interface is thought to be recognized by the E3 ligase Hel2, ZNF598 in mammals, where it ubiquitinates key small subunit r-proteins uS3 and uS10 in yeast, and eS10 and uS10 in mammals (Garzia et al., 2017; Matsuo et al., 2017; Simms, Yan and Zaher, 2017; Sundaramoorthy et al., 2017; Juszkiewicz et al., 2018). Importantly, the cryoEM structure of the collided ribosome reveals how these key ribosomal proteins (r-proteins), along with another r-protein ASC1 (RACK1 in mammals) previously implicated in QC (Kuroha et al., 2010; Letzring et al., 2013), form a composite binding interface that is likely to recruit downstream QC factors with ubiquitin-binding domains. While these ubiquitination events are widely considered to be critical to the subsequent recruitment of downstream QC factors, the interaction between Hel2 and the colliding ribosomes has yet to be captured. In the following sections we will discuss how following the identification of problematic mRNAs, the cell initiates multiple downstream events including mRNA decay, ribosome rescue coupled with targeting of the nascent peptide for proteolytic decay, and, as more recently discovered, translational repression.
mRNA decay and translational repression of mRNAs with ORF-internal stalls
Messenger RNAs containing a stall in the middle of an ORF are targeted for decay in a process broadly referred to as No Go Decay (or NGD). The earliest studies argued that the predominant pathway for NGD involved an endonucleolytic mRNA cleavage and subsequent processing of the decay intermediates by Xrn1 and the exosome (Doma and Parker, 2006). Recent studies from our own group (D’Orazio et al., 2019) provided substantial insights into this process of mRNA decay in yeast. First, D’Orazio et al. used a genetic screen to identify the key endonuclease involved in NGD, Cue2, a low abundance protein with four N-terminal ubiquitin binding sites and a C-terminal hydrolase domain that is part of a family of hydrolase proteins called Small MutS Related, or SMR, proteins. Ribosome profiling data established that Cue2 cleavage activity at internal ORF stalls depends on Hel2 (consistent with work from Ikeuchi et. al) and specifically targets mRNA in the A site of the rotated collided ribosome (D’Orazio et al., 2019; Ikeuchi et al., 2019). Interestingly, SMR domains are structurally related to the C-terminal domain of the bacterial IF3 factor that binds in the A site of bacterial ribosomes (Hussain et al., 2016), agreeing with the predicted placement of the Cue2 SMR domain based on ribosome profiling data. Cue2 has homologs in multicellular organisms such as NONU1 in C. elegans and N4BP2 in mammalian cells (D’Orazio et al., 2019; Glover et al., 2020). As described by Glover et al., these homologs contain a polynucleotide kinase (PNK) domain thought to be involved in phosphorylating cleaved mRNAs for further processing by exonucleases. Cue2 lacks such a domain, but another protein in yeast, Tlr1, contains a PNK domain that is required to phosphorylate the 5’OH of cleaved mRNAs from NGD substrates for further processing (Navickas et al., 2020). While N4BP2 has not yet been implicated in mRNA turnover in mammalian cells, mass spectrometry studies provide evidence that N4BP2 physically associates with colliding ribosomes (Sinha et al., 2020).
Despite strong evidence that Cue2 can target problematic mRNAs for endonucleolytic cleavage in yeast, our studies found that in WT cells, decay of problematic mRNAs occurs predominantly through pathways dependent on Xrn1, and, importantly, not in a manner dependent on Cue2 or the exosome (D’Orazio et al., 2019). These data suggest that problematic mRNAs normally trigger what seems on the surface like “canonical” mRNA decay, though a role for deadenylation has not been established. While Not5 has been implicated in triggering mRNA decay on slowly elongating ribosomes (as discussed above), we speculate that Not5 can not access an empty E site on collided ribosomes since the leading ribosome E site is sterically hindered by the lagging ribosome, which would prevent recruitment of the large CCR4-NOT complex, and the lagging ribosome is in a rotated state where the E site is occupied by tRNA (Juszkiewicz et al., 2018; Ikeuchi et al., 2019; Buschauer et al., 2020). Thus, alternative modes of recruitment of decapping machinery on mRNAs with strong ribosome stalls may need to be employed before exonucleolytic decay from the 5’ end could be initiated. Despite these structural views, it will be important to test whether the Not5-mediated decay pathway contributes to RNA stability on such problematic mRNAs. While it is tempting to speculate that ribosome collisions provide a distinct way for cells to regulate the levels of normal cellular mRNA through Hel2-mediated events, gene-specific studies, RNA-Seq and ribosome profiling data point to very few endogenous substrates whose levels are affected by HEL2 deletion (Sitron, Park and Brandman, 2017; D’Orazio et al., 2019; Matsuo et al., 2020).
Another piece of the QC puzzle has been recently revealed in genetic and proteomic screens searching for factors involved in regulating reporter expression from problematic mRNAs. In a mammalian CRISPR screen, a known translational repressor, GIGYF2 (Morita et al., 2012), along with its binding partner the eIF4E homolog 4E2, was identified as increasing overall expression of the reporter gene when deleted (Hickey et al., 2020). In similar yeast screens, the homologs of GIGYF2, SYH1, and a paralogous gene SMY2, were identified as increasing expression of ORF-stalling reporter gene GFP when deleted (Hickey et al., 2020). And, in two separate proteomic studies in mammalian cells, an adaptor protein that binds to colliding ribosomes, EDF1, was identified, and was shown to be critical for recruiting GIGYF2:4E2 to these complexes (Juszkiewicz, Slodkowicz, et al., 2020; Sinha et al., 2020).
While it was clear from these many studies that problematic mRNAs are specifically targeted by these factors, it was critical in each case to establish whether these factors play a role in translational repression, mRNA decay, or both. GIGYF2 has been previously implicated in translational repression through recruitment of the alternative cap binding factor 4E2 and competition for productive m7G cap binding (Morita et al., 2012); interestingly, there is no known homolog of 4E2 in yeast and so fundamental aspects of regulation may differ in these systems.
Each of the mammalian studies established a clear role for GIGYF2/4E2 in translational repression on the problematic mRNAs tested with no evidence for a role in mRNA degradation (Hickey et al., 2020; Juszkiewicz, Slodkowicz, et al., 2020; Sinha et al., 2020); however, a recent genome wide analysis of GIGYF2 protein function argues for a broader role for these proteins in mediating mRNA decay (Weber et al., 2020). Further targeted experiments will be required to sort through some of the differences and specificities defined in these many studies.
By contrast, deletion of SYH1 and SMY2 in yeast almost fully restores mRNA levels for a problematic mRNA reporter (Hickey et al., 2020). One plausible explanation is that Syh1 and Smy2 promote translational repression, and that this in turn leads directly to mRNA decay, consistent with the known tight coupling of these events throughout biology (Hu et al., 2009; Roy and Jacobson, 2013); alternatively, it is possible that Syh1 and Smy2 function somewhat differently in yeast and directly communicate with the mRNA decay machinery independent of cap binding and translational repression. Future experiments that define how problematic mRNAs are signaled for decapping will possibly reconcile some of these apparent differences between mammalian cells and yeast.
Rescue of ribosomes on mRNAs with ORF-internal stalls
Another important component in ribosome-mediated QC involves the rescue of stalled, trapped ribosomes. Given that stalled ribosomes may never encounter a normal termination signal, preventing access to the normal processes of peptide release (termination) and ribosome dissociation (recycling), the cell has evolved mechanisms to promote these steps as well on problematic mRNAs. Importantly, ribosome rescue is coupled to the targeting of the nascent polypeptide for proteolytic decay.
Early studies implicated Dom34 (PELO in mammals) and its GTPase partner Hbs1 (HBS1L in mammals) in NGD based on the their impact on endonucleolytic cleavage events as revealed by northern blotting of stall inducing mRNAs (Doma and Parker, 2006). Subsequent studies established that Dom34:Hbs1 is not critical for endonucleolytic cleavage per se and noted the structural similarity between Dom34/Hbs1 and eRF1/eRF3 (Dario O. Passos,* Meenakshi K. Doma et al., 2010). In vitro biochemical studies provided the first evidence that Dom34:Hbs1 functions to destabilize the subunit interface to promote dissociation (Shoemaker, Eyler and Green, 2010; Pisareva et al., 2011). Subsequent biochemical studies showed that Rli1 (ABCE1 in mammals), a previously known factor implicated in translation initiation (Dong et al., 2004; Andersen and Leevers, 2007), works together with Dom34 (and its homolog eRF1) to recycle ribosomes in an ATPase-dependent manner, splitting the 80S ribosome into 40S and 60S subunits with the peptidyl-tRNA remaining associated with the 60S subunit (Pisarev et al., 2010; Barthelme et al., 2011; Shoemaker and Green, 2011). Specificity in these Dom34-promoted dissociation reactions was seen for ribosomes positioned on 3’ terminally truncated mRNAs (Shoemaker, Eyler and Green, 2010; Pisareva et al., 2011). Simultaneous cryoEM studies provided structural insights into these recycling/rescue reactions catalyzed by eRF1:Rli1 and Dom34:Rli1 (Becker et al., 2011), and in particular the preferential targeting of ribosomes on shortened mRNAs could be immediately rationalized by the binding of the N-terminal domain of Hbs1 in the mRNA channel (Becker et al., 2011).
Ribosome profiling studies were broadly consistent with these biochemical and structural observations, identifying the predominant in vivo targets of Dom34:Hbs1 as 16 nt long ribosome protected fragments (RPFs), indicative of ribosomes stalled at the end of truncated mRNAs, with an empty A site (Guydosh and Green, 2014, 2017). Similar results were obtained in ribosome profiling studies from C. elegans where they identified short RPFs (16 nts) as abundant ribosome rescue targets for the Dom34 homolog PELOTA (Arribere and Fire, 2018).
These data together established the biochemical specificities of the Dom34:Hbs1 rescue factors in vivo. It was further speculated that these truncated mRNA substrates represent the incomplete products of exonucleolytic decay by the exosome, or in light of new studies, the endonucleolytic cleavage fragments generated by Cue2 or other endonucleases such as Smg6 involved in NMD (Arribere and Fire, 2018; D’Orazio et al., 2019). Consistent with these views, ribosome profiling studies identified endogenous mRNA targets and found them to be enriched in species known to be subject to endonuclease-driven NGD, including prematurely polyadenylated mRNAs, as well as the endonucleolytic cleavage fragments of Ire1 in S. pombe and in S. cerevisiae (Guydosh and Green, 2014, 2017; Guydosh et al., 2017). Importantly, however, we note that Dom34 substrates were most robustly detected in SKI2-deletion backgrounds that artificially stabilized the relatively rare decay intermediates in the cell, thus likely skewing (and potentially over-estimating) the contributions of both endonucleolytic cleavage and Dom34:Hbs1-dependent rescue in these cells.
Despite these insights into Dom34:Hbs1 function and specificity, in vivo genetic results show that, along with Cue2-mediated endonucleolytic cleavage, Dom34:Hbs1 rescue likely represents a minor pathway on problematic mRNAs with ORF-internal stalling sequences in yeast (D’Orazio et al., 2019). Instead, for ORF-triggered collisions, ribosome rescue is thought to be predominantly catalyzed by the yeast Ribosome Quality control Trigger (RQT) complex consisting of the RNA helicase Slh1 (Ski2-like helicase 1) and associated factors Cue3 and Rqt4 (Matsuo et al., 2017, 2020), with homologs in mammalian cells ASCC3, ASCC2 and TRIP4 (the ASC-1 complex), respectively (see Figure 2A) (Matsuo et al., 2017; Juszkiewicz, Speldewinde, et al., 2020). Ribosome profiling experiments performed in yeast carrying stalling reporters reveal an increased abundance of ribosomes around ORF-internal stalling sequences in the absence of Slh1, consistent with the model that Slh1 plays a role in clearing ribosomes by subunit dissociation from these sequences (Sitron, Park and Brandman, 2017; D’Orazio et al., 2019). Biochemical experiments reveal that the substrate for the RQT-dependent splitting reaction is ubiquitinated collided ribosomes, possibly even preferentially the trisome species, and that the splitting reaction depends on the ATPase activity of Slh1 (Juszkiewicz, Speldewinde, et al., 2020; Matsuo et al., 2020). Cue3 possesses ubiquitin-binding domains although the role for these domains in recruitment of RQT to the collided ribosome is inconsistent in yeast and mammals (Matsuo et al., 2017; Juszkiewicz, Speldewinde, et al., 2020). RQT and subsequently the mammalian equivalent, ASCC, were shown to dissociate the lead ribosome in assays that used purified RQT factors (Juszkiewicz, Speldewinde, et al., 2020; Matsuo et al., 2020). Since helicases are known to be processive, it seems possible that the RQT complex iteratively dissociates lead ribosomes along the mRNA from the 3’ end. The ultimate products of the reaction are dissociated large and small subunits, with mRNA bound to the ubiquitylated 40S subunit, and with the peptidyl-tRNA bound to the 60S subunit (Matsuo et al., 2020). This latter complex engages the Ribosome Quality control Complex (RQC) (Brandman et al., 2012; Defenouillere et al., 2013), which ubiquitylates the nascent peptide via Ltn1 leading to degradation by the proteasome (Bengtson and Joazeiro, 2010).
Figure 2:
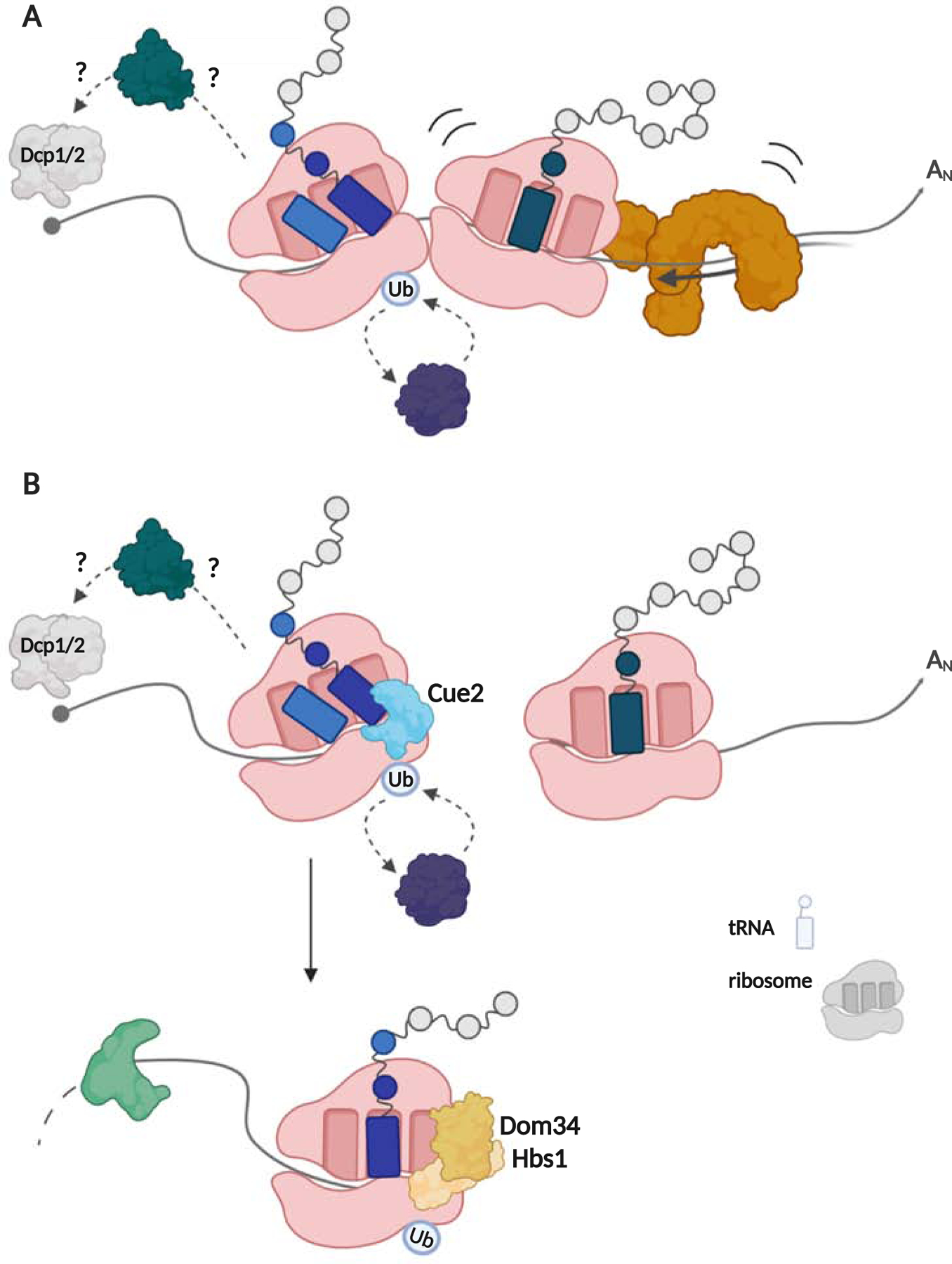
Model for QC on internal ORF-stalled ribosomes. Ribosome collisions signal Hel2-dependent ubiquitination, Syh1-mediated mRNA decay through Xrn1, and both ribosome rescue by RQT (A) and endonucleolytic cleavage by Cue2 followed by Dom34:Hbs1 ribosome rescue (B).
What seems clear from the descriptions of the Dom34:Hbs1 and RQT ribosome rescue systems is that they target different ribosome substrates that can accumulate in the cell. Dom34:Hbs1 targets ribosomes trapped at the end of truncated mRNAs (with no mRNA density in the A site), while RQT targets ribosomes stalled on intact mRNAs. Molecular insights into how these pathways might synergize were revealed in molecular genetic experiments. While it was clear that Cue2-promoted mRNA decay represents a normally minor pathway for ORF-triggered problematic mRNAs (since its deletion did not affect overall reporter mRNA levels), it was found that in cells lacking Slh1, Cue2-promoted mRNA decay became the predominant pathway regulating mRNA abundance (D’Orazio et al., 2019). Given that the RQT clears the stalled colliding ribosomes on ORF-internal problematic mRNAs, these data suggest that when ribosomes accumulate beyond the capacity of this normal rescue pathway, that Cue2-mediated endonucleolytic cleavage provides an alternative route for ribosome rescue, where ribosomes trapped on truncated mRNAs are rescued through the actions of Dom34:Hbs1 (see Figure 2B).
There may be additional contexts in which ribosome stalling occurs without collisions, for example, when initiation rates in the cell or on that mRNA are overall low. These situations may also require ribosome rescue and proteolytic decay to avoid proteotoxic stress. An example of this may be seen in mammalian cells where unresolved ribosome stalling leads to neurodegeneration. In this case, a mammalian homolog of Hbs1, GTPBP2, is required for resolution of ribosomes stalled on Arg codons in mice carrying a mutant brain-specific Arg-tRNA isoform (Ishimura et al., 2014). Analogously, Dom34:Hbs1 may act with some efficiency on a ribosome stalled within an ORF, but the RQT complex may more efficiently act on this complex in the context of a collision.
Taken together, these data argue that at least two pathways are utilized by cells to rescue ribosomes trapped on ORF-internal problematic sequences (and to target their incomplete nascent peptides for decay). The genetic results suggest that while the primary mechanism for QC involves ribosome rescue by the RQT and Xrn1-dependent mechanisms for mRNA decay, endonucleolytic cleavage by Cue2 and ribosome rescue by Dom34:Hbs1 provides a secondary pathway. It is easy to imagine that genetic depletion of Slh1 might mimic a situation where the cell is overwhelmed with problematic mRNAs, and where the normally low abundance QC machinery becomes limiting. For example, general (abundant) damage to the mRNA population in a cell might occur on exposure to oxidative or UV stressors, thus creating a situation where the alternative pathway plays an increasingly essential role.
QC on poly-adenylated mRNAs lacking a stop codon
How the cell identifies mRNAs without a stop codon
Another major class of problematic mRNA in the cell is produced from errors during mRNA processing in the nucleus. After transcription initiation occurs and RNA polymerase is elongating, two key processes – co-transcriptional splicing and transcription termination – must occur to produce a mature, functional transcript. Errors in these processes can lead to a problematic mRNA. For example, errors in splicing most typically result in an mRNA with a premature stop codon that is targeted for Nonsense Mediated Decay (NMD). Errors in transcription termination, however, often produce mRNAs without a stop codon. How does this occur? Transcription elongation is terminated through cleavage of the nascent mRNA transcript from the RNA polymerase followed by polyadenylation of the transcript by poly-A polymerase (Mischo and Proudfoot, 2013). Cleavage and polyadenylation depend on recognition of an A-rich nucleotide stretch in the emerging transcript; however, since recognition of this sequence is not 100% accurate and since there are many near-cognate sites, it is not uncommon for polyadenylation to occur before a stop codon is transcribed, thus disrupting the ORF with poly-A sequence (Mischo and Proudfoot, 2013). Interestingly, disruption of U1 snRNP function dramatically increases premature polyadenylation rates in the cell, and may mimic changes like those observed in cancer cells (Berg et al., 2012; Oh et al., 2017).
When a ribosome translates an mRNA without an in-frame stop codon, it encounters poly-A sequence which encodes iterated lysine residues (encoded by AAA). These mRNAs are referred to as “non-stop” and have been reported to be targeted by QC events including ribosome rescue, translational repression and mRNA and nascent peptide decay (Frischmeyer et al., 2002; Inada and Aiba, 2005). While these QC events sound generally comparable to what has been observed for ORF-internal problematic mRNAs, the molecular details differ, as we describe below.
While it makes intuitive sense that poly-A sequences are recognized as aberrant, how such sequences are recognized to signal QC was not immediately clear. Insights into answering this question have come from biochemical and structural studies. Early studies argued that iterated poly-basic sequences interact with the negatively charged exit tunnel of the ribosome, causing ribosome stalling, and triggering of QC (Ito-Harashima et al., 2007; Lu, Kobertz and Deutsch, 2007). Subsequent studies, however, provided a more nuanced view. First, using bacterial and yeast in vitro reconstituted translation systems, it was reported that ribosomes exhibit large kinetic rate defects in elongation on encountering iterated AAA codons and even slide on poly-A sequences (Chen et al., 2014; Koutmou et al., 2015). Subsequent studies in mammalian cells showed that frameshifting (sliding) events occur in vivo on poly-A sequences within the ORF (Arthur et al., 2015). Such sliding typically results in the ribosome encountering an out of frame premature termination codon and targeting these mRNAs for NMD (Arthur et al., 2015; Koutmou et al., 2015). These studies in bacteria, yeast and mammalian cells also established that poly-Lys encoded by AAG codons is decidedly less problematic for the ribosome. Indeed, iterated lysine sequences encoded for by poly-A stretches are extremely rare within the transcriptome – while there are mRNAs that encode relatively long lysine stretches, in those sequences lysines are disproportionately encoded by AAG relative to AAA (Arthur et al., 2015; Koutmou et al., 2015). Why then do ribosomes struggle to translate iterated lysines encoded for by AAA codons but not by AAG codons?
A series of cryoEM experiments provided detailed structural insight into these stalling events. These structures described an unusual structure that poly-A sequence assumes on its own (Tang et al., 2019) and within the mRNA channel which precludes productive aminoacyl-tRNA binding and readily rationalizes the previously documented kinetic defects in elongation (Chandrasekaran et al., 2019; Tesina, Lessen, et al., 2019). In these structures poly-A sequences are found in a helical conformation in which the backbone of the nucleotide stretch is exposed, while the bases are protected within the helix and are seemingly inaccessible for decoding; 18S rRNA bases (A1756 and C1634 in yeast and A1825 and C1698 in mammalian cells) in the decoding center stack on the single-stranded poly-A helix and stabilize this unusual conformation (Chandrasekaran et al., 2019; Tesina, Lessen, et al., 2019). In addition to revealing the mechanism through which poly-A promotes ribosome frameshifting and impedes translation, the cryoEM studies reveal that poly-A-induced pausing within an ORF leads to ribosome collisions. Interestingly, however, these collided ribosome structures differ somewhat from those associated with CGA-stalling sequences in yeast in that the trailing ribosome assumes a non-rotated state following minor slippage on the homo-polymeric template.
It is not clear what the key signaling event is for QC on ribosomes stalled within a poly-A tail. Is the signaling event ribosome collisions, the aberrant structure of the poly-A tail within the mRNA channel, or possibly the positioning of the ribosome in close proximity to the 3’ end of the mRNA? As we will discuss, the factors involved in QC on non-stop mRNAs are distinct from those that act on ribosomes stalled on an ORF-internal poly-A sequence. While many of these factors have long been known, recent advances are bringing clarity to the molecular mechanisms of these processes.
mRNA decay and ribosome rescue are closely linked on non-stop mRNAs
Degradation of non-stop mRNAs is strongly dependent on the SKI complex (SKI2, SKI3, SKI8, and the exosome adapter protein SKI7); levels of mRNAs lacking a stop codon are nearly restored on deletion of the SKI complex, but are unaffected by deletion of XRN1 (van Hoof et al., 2002). Meanwhile, the levels of mRNAs containing ORF-internal stalls are unaffected on deletion of the SKI complex, but are restored on deletion of XRN1 (D’Orazio et al., 2019). Therefore, ribosomes stalled within a poly-A tail and ribosomes stalled within an open reading frame are clearly distinguished from one another in the cell on the basis of their decay by orthogonal decay machineries.
Genetic evidence thus points to the possibility that the SKI complex plays a role in recognition of stalled ribosomes in the poly-A tail. Structural studies reveal how the SKI complex interacts with a ribosome with a short 3’ mRNA extending from the mRNA channel, directly binding to both the 3’ end of the mRNA and the 40S subunit of the ribosome; indeed, recruitment of the SKI complex in vitro is greatly increased by RNase digestion that shortens the mRNA 3’ end tail (Schmidt et al., 2016). These observations suggest a possible model wherein a stalled ribosome at or near the end of an mRNA directly recruits the SKI complex, allowing the mRNA to be funneled into the exosome through the helicase module, Ski2. Whether or how the ribosome clears the poly-A binding protein (PABP) from the poly-A tail making it accessible to the SKI complex is not known.
Interestingly, recent biochemical data provide evidence for a role for the SKI complex in ribosome rescue. Using toe-printing assays and an in vitro reconstituted mammalian translation system, Zinoviev et al. demonstrate that the SKI complex is able to effectively pull mRNA from a translating ribosome until the mRNA dissociates from the ribosome entirely (Zinoviev et al., 2020). At this stage, the 80S ribosome still likely contains the peptidyl-tRNA and may serve as a substrate for Dom34:Hbs1, somewhat akin to ribosomes trapped on truncated mRNA derived from Cue2 cleavage (see Figure 3). Importantly, the SKI complex includes the exosome binding factor SKI7, such that as the SKI complex pulls the mRNA from the ribosome, it can thread it directly into the exosome for degradation (Halbach et al., 2013).
Figure 3:
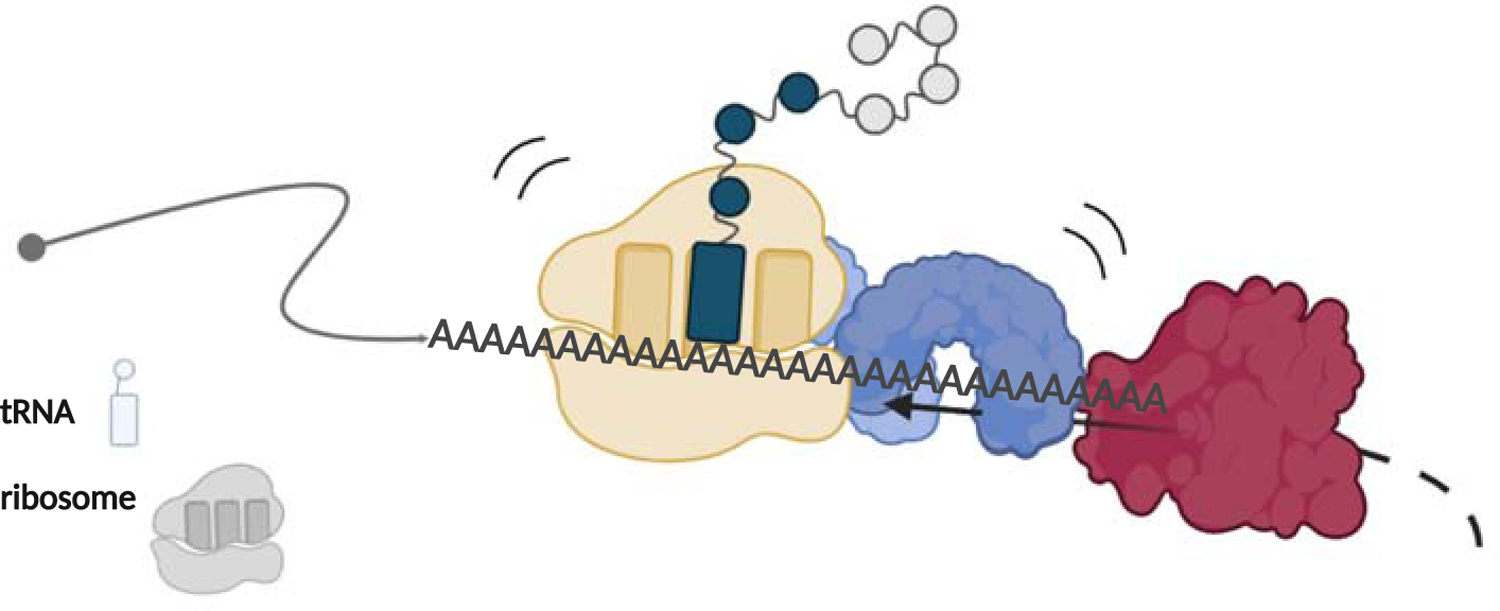
Model for QC on poly-A stalled ribosomes. Ribosome stalling on poly-A sequences recruits the SKI complex through an unknown mechanism. The SKI complex then “pulls” on the mRNA from the ribosome and funnels it into the exosome.
Figures created with BioRender.com
An interesting parallel is that the helicase in the SKI complex, Ski2, is highly related to the ribosome rescue factor, Slh1, thought to function on ORF-internal stalled ribosomes. Ski2 and Slh1 are both RNA dependent helicases, exhibit substantial homology to one another, and were initially implicated in combatting viruses in yeast (Martegani et al., 1997). Due to these similarities, one might speculate that the mechanism of the RQT complex is similar to that of the SKI complex, involving an iterative removal of ribosomes through the action of the helicase domain. In vitro data for yeast and mammalian RQT reveal a dependence on the ATPase function of Slh1/ASCC3 for ribosome clearing, though the processivity of this complex was not established (Matsuo et al., 2017, 2020; Juszkiewicz, Speldewinde, et al., 2020). Direct comparisons of these complexes and their activities on common substrates would likely be helpful in sorting through these potential similarities and differences.
Distinct from the RQT complex, the SKI complex is bound by the exosome, the major 3’ to 5’ RNA decay machine, thus allowing for direct coupling of two quality control events, mRNA decay and ribosome rescue. Indeed, genetic studies show that the protein product from a non-stop message and the mRNA itself are both stabilized upon deletion of the SKI complex, confirming that the SKI complex reaction doubles as a facilitator of ribosome rescue and mRNA decay. In contrast, the RQT complex leaves behind an intact mRNA that is ultimately degraded by Xrn1. We note that the coupling of RQT activity to mRNA decay in many cases has been difficult to detect, although genetic screens and proteomics approaches suggest decay factors are functioning and physically present at ribosome stalling sites (Hickey et al., 2020; Sinha et al., 2020). From an evolutionary perspective, it is interesting to consider why mRNA decay and ribosome rescue might be more tightly coupled on prematurely polyadenylated mRNAs than on damaged or stall-containing mRNAs.
Degradation of truncated mRNAs
The SKI complex also assists in degradation of cytoplasmic mRNAs wholly lacking a poly-A tail – this molecular species may be generated by endonucleolytic (e.g. Ire1 or Cue2) or exonucleolytic processing of mRNAs. As mentioned earlier, when a ribosome encounters the 3’ end of this tail-less mRNA, Dom34:Hbs1 is thought to recognize the empty A site and catalyze ribosome subunit dissociation, or ribosome rescue. This now liberated 3’ end, lacking both a poly-A tail and a ribosome, likely serves as a good substrate for the SKI complex and progressive mRNA degradation (Schmidt et al., 2016). For such mRNAs, we suspect that this cooperation between Dom34:Hbs1 and the SKI complex effectively drives mRNA decay and ribosome rescue to completion. The peptidyl-tRNAs associated with the 60S subunit are thought to be processed by the RQC, just like the equivalent complexes generated by RQT-dependent splitting. We see in this example how multiple systems for clearing ribosomes, SKI and Dom34:Hbs1, collaborate to bring about QC and thus reveal the resilience of these pathways in managing problematic mRNAs in the cell.
Conclusions
What we have described throughout this review is how cells utilize multiple pathways to regulate output from problematic mRNAs in the cell. Each pathway targets stalled ribosomes, through machinery that has likely evolved to maximize efficiency of stall resolution and to prevent the build up of toxic peptides, but with somewhat differing specificities. For example, the cell does not recruit the SKI complex to an ORF-internal stall because the SKI complex has evolved to engage free 3’ ends of mRNAs. Instead, the cell recruits the RQT complex, which latches on to the mRNA through an unknown mechanism to rescue the internally-stalled ribosomes. ORF-internal stalled, collided ribosomes signal mRNA decay through an unknown mechanism that ultimately triggers Xrn1-mediated decay from the 5’ end of the mRNA. If the cell cannot resolve this ORF-internal stall with the RQT complex, an endonuclease Cue2 is recruited to cleave the mRNA, leading to ribosome rescue and mRNA decay through alternate pathways involving Dom34:Hbs1 and the SKI complex (see Figure 2). Meanwhile, if a ribosome is stuck on a poly-A tail, the cell does not recruit Cue2 or Xrn1, but instead the SKI complex is essential for both degradation and ribosome rescue (see Figure 3).
While these pathways exhibit some specificity, they clearly collaborate to rid the cell of dangerous mRNA species. This was first noted in early studies where both slh1Δ and ski2Δ cells were sensitive to non-poly-adenylated viral RNA, and the slh1Δski2Δ strain showed an increased sensitivity (Searfoss and Wickner, 2000). However, many genetic experiments were confounded by redundancy since deletion of components from one pathway ultimately allowed another pathway to compensate. For example, when the SKI complex is deleted and can not resolve ribosome complexes trapped on a truncated mRNA, Xrn1, Cue2 and Dom34:Hbs1 come to the rescue (Ikeuchi and Inada, 2016). Or, in another example, while nonsense mediated decay (NMD) is mediated by quite a distinct set of molecular players from those we have discussed here, recent studies have shown that the NMD pathway converges with the NGD pathways for completion of mRNA degradation (Arribere and Fire, 2018). It turns out that a key early step in NMD is cleavage of the PTC-containing mRNA with an endonuclease, SMG6, which leads to the accumulation of ribosomes on truncated mRNAs, and the need for processing by Pelota:HBS1l (Dom34:Hbs1), the SKI complex, and likely the metazoan homolog of Cue2, N4BP2. In a final example, the balance between Xrn1- and Cue2- dependent RNA decay as well as RQT- and Dom34:Hbs1-dependent ribosome rescue on internal ORF stalling sequences is likely determined by the relative abundance of collisions and of QC machinery – when one pathway fails, another compensates (D’Orazio et al., 2019).
The redundancy of these pathways speaks to their importance. As we have outlined here, we have a broad understanding of the ribosome-based triggers of QC, though there remain gaps in our knowledge. We have little biochemical or structural understanding as to how QC factors engage the colliding (or not) ribosomes to trigger the downstream events. There are unknowns surrounding signaling to mRNA decay machinery and translation repression, and we are just beginning to understand how ribosomes are rescued from these problematic sequences. And, while mRNA decay appears to be an important part of QC in yeast, the importance of these pathways is just beginning to be thoroughly examined in mammalian systems. One new frontier will be the exploration of these QC systems in cells or tissues with unusual or demanding protein synthesis regimes, where the limitations and extremes of the systems may be revealed.
D’Orazio and Green analyze ribosome-dependent pathways involved in RNA quality control. They discuss how the many states of a ribosome during translation of a problematic mRNA signal a number mRNA quality control pathways, with overlapping and compensatory mechanisms that ensure mRNA and protein degradation.
Acknowledgements:
We thank members of the Green lab and colleagues for the helpful discussions about the ideas in this review. This material is based upon work supported by grants from the National Institutes of Health (NIH), R37GM059425 to the lab of Rachel Green and 5T32GM007445-39 to the Johns Hopkins University School of Medicine graduate program BCMB. Any opinions and interpretations in this review are those of the authors and do not necessarily reflect the views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Rachel Green is a member of the scientific advisory board at the journal Molecular Cell.
References
- Andersen DS and Leevers SJ (2007) ‘The essential drosophila ATP-binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation’, Journal of Biological Chemistry, 282(20), pp. 14752–14760. doi: 10.1074/jbc.M701361200. [DOI] [PubMed] [Google Scholar]
- Anderson JSJ and Parker RP (1998) ‘The 3’ to 5’ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3’ to 5’ exonucleases of the exosome complex.’, The EMBO journal, 17(5), pp. 1497–506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA and Fire AZ (2018) ‘Nonsense mRNA suppression via nonstop decay’, eLIFE, 7, pp. 1–23. doi: 10.7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P and Djuranovic S (2015) ‘Translational control by lysine-encoding A-rich sequences.’, Science advances, 1(6). doi: 10.1126/sciadv.1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers S-V, Londei P, Ermler U and Tampé R (2011) ‘Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1.’, Proceedings of the National Academy of Sciences of the United States of America, 108(8), pp. 3228–33. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache J-P, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O and Beckmann R (2011) ‘Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome.’, Nature structural & molecular biology, 18(6), pp. 715–20. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W and Rodnina MV (2016) ‘Choreography of molecular movements during ribosome progression along mRNA’, Nature Structural and Molecular Biology, 23(4), pp. 342–348. doi: 10.1038/nsmb.3193. [DOI] [PubMed] [Google Scholar]
- Bengtson MH and Joazeiro CAP (2010) ‘Role of a ribosome-associated E3 ubiquitin ligase in protein quality control.’, Nature, 467(7314), pp. 470–3. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L et al. (2012) ‘U1 snRNP determines mRNA length and regulates isoform expression’, Cell, 150(1), pp. 53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boël G, Letso R, Neely H, Price WN, Wong KH, Su M, Luff JD, Valecha M, Everett JK, Acton TB et al. (2016) ‘Codon influence on protein expression in E. coli correlates with mRNA levels’, Nature, 529(7586), pp. 358–363. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W, Zhou S, King D, Shen PS, Weibezahn J et al. (2012) ‘A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress.’, Cell, 151(5), pp. 1042–54. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O and Hegde RS (2016) ‘Ribosome-associated protein quality control.’, Nature structural & molecular biology, 23(1), pp. 7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschauer R, Matsuo Y, Sugiyama T, Chen YH, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Nobuta R et al. (2020) ‘The Ccr4-Not complex monitors the translating ribosome for codon optimality’, Science, 368(6488). doi: 10.1126/science.aay6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Muhlrad D and Parker R (1993) ‘A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons.’, Molecular and Cellular Biology, 13(9), pp. 5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LY, Mugler CF, Heinrich S, Vallotton P and Weis K (2018) ‘Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability’, eLife, 7, pp. 1–32. doi: 10.7554/eLife.32536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran V, Juszkiewicz S, Choi J, Puglisi JD, Brown A, Shao S, Ramakrishnan V and Hegde RS (2019) ‘Mechanism of ribosome stalling during translation of a poly(A) tail.’, Nature structural & molecular biology, 26(12), pp. 1132–1140. doi: 10.1038/s41594-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CYA and Shyu A. Bin (2011) ‘Mechanisms of deadenylation-dependent decay’, Wiley Interdisciplinary Reviews: RNA, 2(2), pp. 167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Johansson M, Tsai A, O’Leary SE and Puglisi JD (2014) ‘Dynamic pathways of −1 translational frameshifting.’, Nature, 512(7514), pp. 328–32. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA and Weiss B (2020) ‘Ribosome pausing, a dangerous necessity for co-translational events’, Nucleic acids research, 48(3), pp. 1043–1055. doi: 10.1093/nar/gkz763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio KN, Wu CC-C, Sinha N, Loll-Krippleber R, Brown GW and Green R (2019) ‘The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay.’, eLife, 8, pp. 1–27. doi: 10.7554/eLife.49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dario O. Passos,* Meenakshi K. Doma, †, Shoemaker CJ, Denise Muhlrad RG, Weissman J, Julie Hollien,? and Parker*, and R. (2010) ‘Analysis of Dom34 and Its Function in No-Go Decay’, Molecular biology of the cell, 21(22), pp. 4042–4056. doi: 10.1091/mbc.E09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A et al. (2013) ‘Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products’, Proceedings of the National Academy of Sciences, 110(13), pp. 5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Copeland BR, Mustoe AM and Goldstein DB (2020) ‘Natural Selection Shapes Codon Usage in the Human Genome.’, American journal of human genetics, 107(1), pp. 83–95. doi: 10.1016/j.ajhg.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK and Parker R (2006) ‘Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation’, Nature, 440(7083), pp. 561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H and Hinnebusch AG (2004) ‘The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly.’, The Journal of biological chemistry, 279(40), pp. 42157–68. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- Forrest ME, Pinkard O, Martin S, Sweet TJ, Hanson G and Coller J (2020) ‘Codon and amino acid content are associated with mRNA stability in mammalian cells.’, PloS one, 15(2), p. e0228730. doi: 10.1371/journal.pone.0228730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R and Dietz HC (2002) ‘An mRNA surveillance mechanism that eliminates transcripts lacking termination codons.’, Science (New York, N.Y.), 295(5563), pp. 2258–61. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T et al. (2017) ‘The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs.’, Nature communications, 8(May), p. 16056. doi: 10.1038/ncomms16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover ML, Burroughs AM, Monem PC, Egelhofer TA, Pule MN, Aravind L and Arribere JA (2020) ‘NONU-1 Encodes a Conserved Endonuclease Required for mRNA Translation Surveillance’, Cell Reports, 30(13), pp. 4321–4331.e4. doi: 10.1016/j.celrep.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJE, Satory D, Halliday JA and Herman C (2015) ‘Lost in transcription: Transient errors in information transfer’, Current Opinion in Microbiology, 24, pp. 80–87. doi: 10.1016/j.mib.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh N and Green R (2017) ‘Translation of poly(A) tails leads to precise mRNA cleavage’, Rna, 23, pp. 749–761. doi: 10.1261/rna.060418.116.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Kimmig P, Walter P and Green R (2017) ‘Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe.’, eLife, 6. doi: 10.7554/eLife.29216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR and Green R (2014) ‘Dom34 rescues ribosomes in 3’ untranslated regions.’, Cell, 156(5), pp. 950–62. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Reichelt P, Rode M and Conti E (2013) ‘The yeast ski complex: crystal structure and RNA channeling to the exosome complex.’, Cell, 154(4), pp. 814–26. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Han P, Shichino Y, Schneider-Poetsch T, Mito M, Hashimoto S, Udagawa T, Kohno K, Yoshida M, Mishima Y, Inada T et al. (2020) ‘Genome-wide Survey of Ribosome Collision’, Cell Reports, 31(5). doi: 10.1016/j.celrep.2020.107610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G and Coller J (2017) ‘Codon optimality, bias and usage in translation and mRNA decay’, Nature Reviews Molecular Cell Biology, 19, p. 20. Available at: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey KL, Dickson K, Cogan JZ, Replogle JM, Schoof M, D’Orazio KN, Sinha NK, Hussmann JA, Jost M, Frost A et al. (2020) ‘GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control’, Molecular Cell, pp. 1–13. doi: 10.1016/j.molcel.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A, Kastelein RA, Vasser M and de Boer HA (1987) ‘Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression.’, Molecular and Cellular Biology, 7(8), pp. 2914–2924. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC and Parker R (2002) ‘Exosome-mediated recognition and degradation of mRNAs lacking a termination codon.’, Science (New York, N.Y.), 295(5563), pp. 2262–4. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE and Coller J (2009) ‘Co-translational mRNA decay in Saccharomyces cerevisiae.’, Nature, 461(7261), pp. 225–9. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Wimberly BT, Kieft JS and Ramakrishnan V (2016) ‘Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.’, Cell, 167(1), pp. 133–144.e13. doi: 10.1016/j.cell.2016.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R and Inada T (2019) ‘Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.’, The EMBO journal, 38(5), p. e100276. doi: 10.15252/embj.2018100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K and Inada T (2016) ‘Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA’, Scientific Reports, 6(June), pp. 1–10. doi: 10.1038/srep28234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T (2020) ‘Quality controls induced by aberrant translation’, Nucleic acids research, 48(3), pp. 1084–1096. doi: 10.1093/nar/gkz1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T and Aiba H (2005) ‘Translation of aberrant mRNAs lacking a termination codon or with a shortened 3’UTR is repressed after initiation in yeast’, EMBO Journal, 24(8), pp. 1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang X, Schimmel P, Senju S, Nishimura Y, Chuang JH and Ackerman SL (2014) ‘RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration.’, Science (New York, N.Y.), 345(6195), pp. 455–9. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S, Kuroha K, Tatematsu T and Inada T (2007) ‘Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast.’, Genes & development, 21(5), pp. 519–24. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V and Hegde RS (2018) ‘ZNF598 Is a Quality Control Sensor of Collided Ribosomes’, Molecular Cell, pp. 1–13. doi: 10.1016/j.molcel.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Slodkowicz G, Lin Z, Freire-Pritchett P, Peak-Chew SY and Hegde RS (2020) ‘Ribosome collisions trigger cis-acting feedback inhibition of translation initiation’, eLife, 9, pp. 1–29. doi: 10.7554/eLife.60038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Speldewinde SH, Wan L, Svejstrup JQ and Hegde RS (2020) ‘The ASC-1 Complex Disassembles Collided Ribosomes.’, Molecular cell, in press, pp. 1–12. doi: 10.1016/j.molcel.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S and Green R (2015) ‘Ribosomes slide on lysine-encoding homopolymeric A stretches’, eLife, 2015(4), pp. 1–18. doi: 10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K and Inada T (2010) ‘Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest.’, EMBO reports, 11(12), pp. 956–61. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Wolf AS, Brule CE and Grayhack EJ (2013) ‘Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1’, Rna, 19(9), pp. 1208–1217. doi: 10.1261/rna.039446.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM and Grayhack EJ (2010) ‘Control of translation efficiency in yeast by codon-anticodon interactions.’, RNA (New York, N.Y.), 16(12), pp. 2516–28. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y (2020) ‘A code within the genetic code: codon usage regulates co-translational protein folding.’, Cell communication and signaling : CCS, 18(1), p. 145. doi: 10.1186/s12964-020-00642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Kobertz WR and Deutsch C (2007) ‘Mapping the Electrostatic Potential within the Ribosomal Exit Tunnel’, Journal of Molecular Biology, 371(5), pp. 1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Maraia RJ and Arimbasseri AG (2017) ‘Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon-Codon Use.’, Biomolecules, 7(1). doi: 10.3390/biom7010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martegani E, Vanoni M, Mauri I, Rudoni S, Saliola M and Alberghina L (1997) ‘Identification of gene encoding a putative RNA-helicase, homologous to SKI2, in chromosome VII of Saccharomyces cerevisiae’, Yeast, 13(4), pp. 391–397. doi: . [DOI] [PubMed] [Google Scholar]
- Martin PB, Kigoshi-Tansho Y, Sher RB, Ravenscroft G, Stauffer JE, Kumar R, Yonashiro R, Müller T, Griffith C, Allen W et al. (2020) ‘NEMF mutations that impair ribosome-associated quality control are associated with neuromuscular disease.’, Nature communications, 11(1), p. 4625. doi: 10.1038/s41467-020-18327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K et al. (2017) ‘Ubiquitination of stalled ribosome triggers ribosome-associated quality control.’, Nature communications, 8(1), p. 159. doi: 10.1038/s41467-017-00188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Tesina P, Nakajima S, Mizuno M, Endo A, Buschauer R, Cheng J, Shounai O, Ikeuchi K, Saeki Y et al. (2020) ‘RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1’, Nature Structural and Molecular Biology, 27(4), pp. 323–332. doi: 10.1038/s41594-020-0393-9. [DOI] [PubMed] [Google Scholar]
- Mischo HE and Proudfoot NJ (2013) ‘Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast.’, Biochimica et biophysica acta, 1829(1), pp. 174–85. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y and Tomari Y (2016) ‘Codon Usage and 3’ UTR Length Determine Maternal mRNA Stability in Zebrafish’, Molecular Cell, 61(6), pp. 874–885. doi: 10.1016/j.molcel.2016.02.027. [DOI] [PubMed] [Google Scholar]
- Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC, Alain T, Fonseca BD, Karashchuk G, Bennett CF et al. (2012) ‘A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development.’, Molecular and cellular biology, 32(17), pp. 3585–93. doi: 10.1128/MCB.00455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler CF, Hondele M, Heinrich S, Sachdev R, Vallotton P, Koek AY, Chan LY and Weis K (2016) ‘ATPase activity of the DEAD-box protein Dhh1 controls processing body formation’, eLife, 5(OCTOBER2016). doi: 10.7554/eLife.18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navickas A, Chamois S, Saint-Fort R, Henri J, Torchet C and Benard L (2020) ‘No-Go Decay mRNA cleavage in the ribosome exit tunnel produces 5′-OH ends phosphorylated by Trl1’, Nature Communications, 11(1), pp. 1–11. doi: 10.1038/s41467-019-13991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J-M, Di C, Venters CC, Guo J, Arai C, So BR, Pinto AM, Zhang Z, Wan L, Younis I et al. (2017) ‘U1 snRNP telescripting regulates a size-functionstratified human genome.’, Nature structural & molecular biology, 24(11), pp. 993–999. doi: 10.1038/nsmb.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R (2012) ‘RNA degradation in Saccharomyces cerevisae.’, Genetics, 191(3), pp. 671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W and Steinmetz LM (2015) ‘Widespread Co-translational RNA Decay Reveals Ribosome Dynamics.’, Cell, 161(6), pp. 1400–12. doi: 10.1016/j.cell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CUT and Pestova TV (2010) ‘The Role of ABCE1 in Eukaryotic Posttermination Ribosomal Recycling’, Molecular Cell, 37(2), pp. 196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV and Pisarev AV (2011) ‘Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes.’, The EMBO journal, 30(9), pp. 1804–17. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR et al. (2015) ‘Codon optimality is a major determinant of mRNA stability’, Cell, 160(6), pp. 1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen Y, Martin S, Alhusaini N, Green R and Coller J (2016) ‘The DEAD-Box Protein Dhh1p Couples mRNA Decay and Translation by Monitoring Codon Optimality.’, Cell, 167(1), pp. 122–132.e9. doi: 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan N, Pochopien AA, Chih-Chien Wu C, Beckert B, Blanchet S, Green R, Rodnina MV and Wilson DN (2020) ‘eEF3 promotes late stages of tRNA translocation on the ribosome’, bioRxiv, p. 2020.07.01.182105. doi: 10.1101/2020.07.01.182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R and Wernisch L (2004) ‘Solving the riddle of codon usage preferences: a test for translational selection.’, Nucleic acids research, 32(17), pp. 5036–44. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV (2016) ‘The ribosome in action: Tuning of translational efficiency and protein folding’, Protein Science, 25, pp. 1390–1406. doi: 10.1002/pro.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B and Jacobson A (2013) ‘The intimate relationships of mRNA decay and translation.’, Trends in genetics : TIG, 29(12), pp. 691–9. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Kowalinski E, Shanmuganathan V, Defenouillère Q, Braunger K, Heuer A, Pech M, Namane A, Berninghausen O, Fromont-Racine M et al. (2016) ‘The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex.’, Science (New York, N.Y.), 354(6318), pp. 1431–1433. doi: 10.1126/science.aaf7520. [DOI] [PubMed] [Google Scholar]
- Searfoss AM and Wickner RB (2000) ‘3′ poly(A) is dispensable for translation’, Proceedings of the National Academy of Sciences of the United States of America, 97(16), pp. 9133–9137. doi: 10.1073/pnas.97.16.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE and Green R (2010) ‘Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay.’, Science (New York, N.Y.), 330(6002), pp. 369–72. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ and Green R (2011) ‘Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast.’, Proceedings of the National Academy of Sciences of the United States of America, 108(51), pp. E1392–8. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL and Zaher HS (2017) ‘Ribosome Collision Is Critical for Quality Control during No-Go Decay’, Molecular Cell, 68(2), pp. 361–373.e5. doi: 10.1016/j.molcel.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NK, Ordureau A, Best K, Saba JA, Zinshteyn B, Sundaramoorthy E, Fulzele A, Garshott DM, Denk T, Thoms M et al. (2020) ‘EDF1 coordinates cellular responses to ribosome collisions’, eLife, 9, pp. 1–44. doi: 10.7554/elife.58828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitron CS, Park JH and Brandman O (2017) ‘Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation’, Rna, 23(5), pp. 798–810. doi: 10.1261/rna.060897.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A and Bennett EJ (2017) ‘ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation’, Molecular Cell, 65(4), pp. 751–760.e4. doi: 10.1016/j.molcel.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TTL, Stowell JAW, Hill CH and Passmore LA (2019) ‘The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.’, Nature structural & molecular biology, 26(6), pp. 433–442. doi: 10.1038/s41594-019-0227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y and Akimitsu N (2012) ‘Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals.’, Genome research, 22(5), pp. 947–56. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesina P, Lessen LN, Buschauer R, Cheng J, Wu CC-C, Berninghausen O, Buskirk AR, Becker T, Beckmann R and Green R (2019) ‘Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.’, The EMBO journal, p. e103365. doi: 10.15252/embj.2019103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesina P, Heckel E, Cheng J, Fromont-Racine M, Buschauer R, Kater L, Beatrix B, Berninghausen O, Jacquier A, Becker T et al. (2019) ‘Structure of the 80S ribosome-Xrn1 nuclease complex.’, Nature structural & molecular biology, 26(4), pp. 275–280. doi: 10.1038/s41594-019-0202-5. [DOI] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW and Puglisi JD (2010) ‘Real-time tRNA transit on single translating ribosomes at codon resolution’, Nature, 464(7291), pp. 1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E and Winkler GS (2013) ‘RNA decay machines: Deadenylation by the Ccr4-Not and Pan2-Pan3 complexes’, Biochimica et Biophysica Acta - Gene Regulatory Mechanisms, 1829(6–7), pp. 561–570. doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Weber R, Chung M-Y, Zinnall U, Landthaler M, Valkov E, Izaurralde E and Igreja C (2020) ‘4EHP and GIGYF1/2 Mediate Translation-Coupled Messenger RNA Decay’, SSRN Electronic Journal. doi: 10.2139/ssrn.3548260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MW, Chen YH, Stowell JAW, Alhusaini N, Sweet T, Graveley BR, Coller J and Passmore LA (2018) ‘mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases’, Molecular Cell, 70(6), pp. 1089–1100.e8. doi: 10.1016/j.molcel.2018.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MW, Stowell JA and Passmore LA (2019) ‘RNA-binding proteins distinguish between similar sequence motifs to promote targeted deadenylation by Ccr4-Not’, eLife, 8, pp. 1–26. doi: 10.7554/eLife.40670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CCC, Peterson A, Zinshteyn B, Regot S and Green R (2020) ‘Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate’, Cell, 182(2), pp. 404–416.e14. doi: 10.1016/j.cell.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, Wright M and Bazzini AA (2019) ‘Translation affects mRNA stability in a codon-dependent manner in human cells.’, eLife, 8, pp. 1–22. doi: 10.7554/eLife.45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LL, Simms CL, McLoughlin F, Vierstra RD and Zaher HS (2019) ‘Oxidation and alkylation stresses activate ribosome-quality control’, Nature Communications, 10(1), pp. 1–15. doi: 10.1038/s41467-019-13579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LL and Zaher HS (2019) ‘How do cells cope with RNA damage and its consequences?’, The Journal of biological chemistry, 294(41), pp. 15158–15171. doi: 10.1074/jbc.REV119.006513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD and Tanenbaum ME (2016) ‘Dynamics of Translation of Single mRNA Molecules in Vivo’, Cell, 165(4), pp. 976–989. doi: 10.1016/j.cell.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Park J, Ha M, Lim J, Chang H and Kim VN (2018) ‘PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay’, Molecular Cell, 70(6), pp. 1081–1088.e5. doi: 10.1016/j.molcel.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS and Liu Y (2015) ‘Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding’, Molecular Cell, 59(5), pp. 744–754. doi: 10.1016/j.molcel.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Ayupov RK, Abaeva IS, Hellen CUT and Pestova TV (2020) ‘Extraction of mRNA from Stalled Ribosomes by the Ski Complex.’, Molecular cell, 77(6), pp. 1340–1349.e6. doi: 10.1016/j.molcel.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


