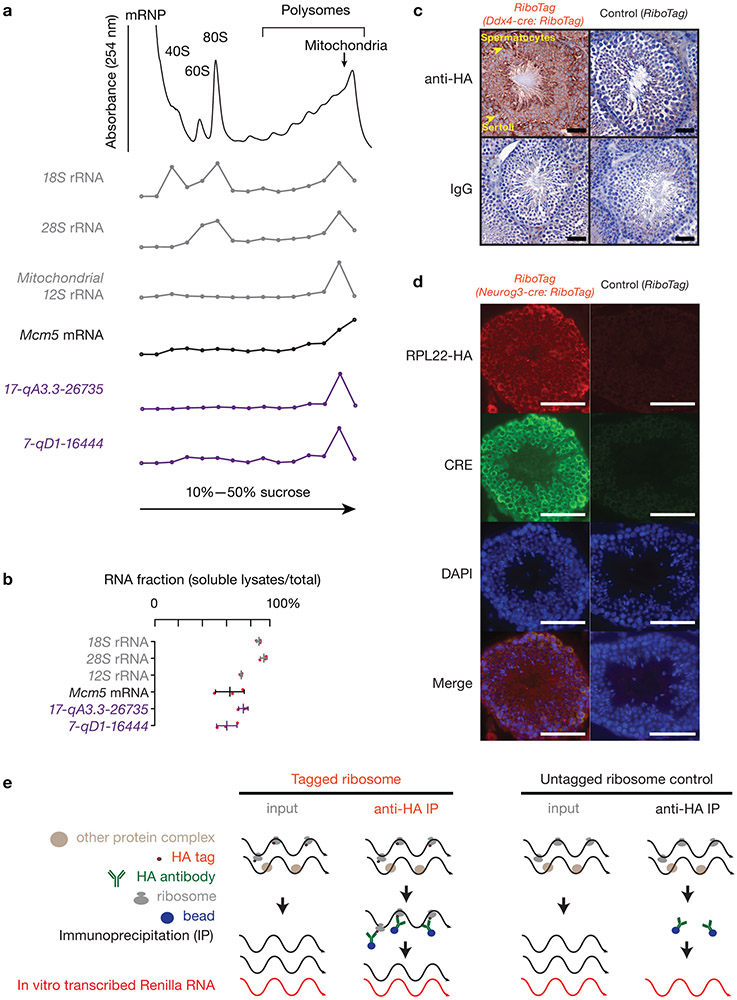
Extended Data Fig. 2. Ribosomes associate with pachytene piRNA precursors.
a, A254 absorbance profile of testis lysates from adult mice following separation in 10% to 50% sucrose density gradients. The testes were lysed in amended lysis buffer that preserves mitochondria. Top to bottom, relative abundance of 18S rRNA, 28S rRNA, 12S rRNA, Mcm5 mRNA, and two pachytene piRNA precursors: 17-qA3.3-26735 and 7-qD1-16444. For Extended Data Figure 2 a, c, d, experiments have been repeated for at least three times independently with similar results. b, Fraction of RNAs in lysates relative to total RNAs in both lysates and pellets. The long transcripts were quantified using RT-qPCR (three independent biological samples; mean ± standard deviation, n = 3 independent experiments). Quantification of piRNA precursors in pellets during lysate preparation revealed that 63 ± 9% of 7-qD1-16444 and 77 ± 4% of 17-qA3.3-26735 precursors were present in soluble lysates, as were 65 ± 13% of Mcm5 mRNA and 75 ± 1% of 12S rRNA. Each data point was overlaid as dot plots. c, Immunohistochemical staining on 6-μm testis sections from Ddx4-cre:RiboTag mice (left 2 panels) and control littermates (without Ddx4-cre, right 2 panels) using anti-HA (upper 2 panels) and mouse IgG isotype control (bottom 2 panels). HA expression in Ddx4-cre:RiboTag mice (upper left) was most prominent in the cytosol of spermatocytes (marked with arrow), but not in Sertoli cells (marked with arrow). No staining was seen in RiboTag control testis (upper right) or sections probed using mouse IgG isotype control as the primary antibody (bottom two panels). Scale bar, 50 μm. d, Immunostaining of testis cryosections from Neurog3-cre:RiboTag mice (left) and from control littermates (without Neurog3-cre, right) using anti-HA, anti-CRE, DAPI, and merged (top to bottom). Scale bar, 75 μm. e, Schematic of affinity purification procedure for ribosome-associated RNAs. In vitro transcribed Renilla RNA was used as a spike-in for normalization. Statistical Source Data are provided in Source Data Extended Data Figure 2.