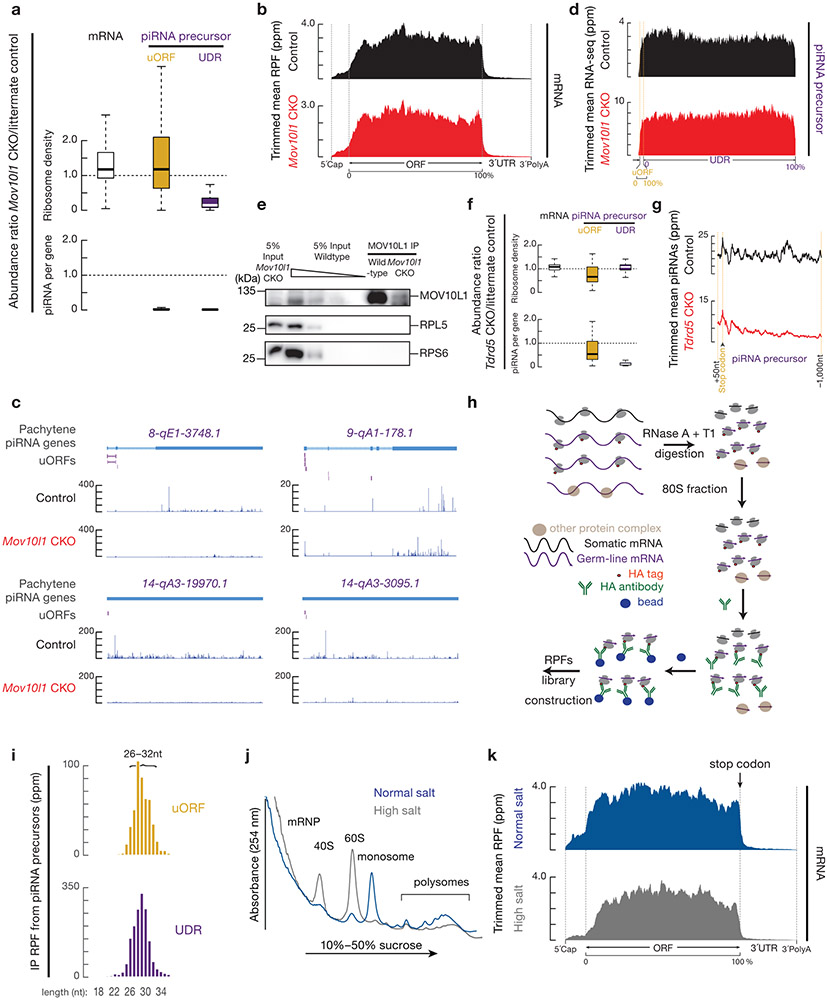
Extended Data Fig. 6. UDR RPFs are bona fide ribosome footprints.
a, Boxplots of the change in ribosome density (upper) or piRNA abundance (lower) per gene in Mov10l1 mutants (Mov10l1CKO/Δ Neurog3-cre, lower) compared to littermate controls (Mov10l1CKO/Δ, upper) in testes. Experiments in a and f have been repeated for three biological replicates and mean is used for plotting. Sample sizes in a and f, n = 100 pachytene piRNA precursors; n =115 pachytene expressed mRNAs. b, Aggregated data for RPF abundance on mRNAs (10% trimmed mean). c, Browser views of piRNA precursor loci with normalized degradome reads. No degradome peak around stop codon was observed. d, Aggregated data for RNA-seq abundance on pachytene piRNA precursors (10% trimmed mean). e, Western blot analyses of RPL5 (Ribosomal large subunit component), RPS6 (Ribosomal small subunit component), and MOV10L1. Experiments have been repeated for two times independently with similar results. f, Boxplots of the change in ribosome density (upper) or piRNA abundance (lower) per gene in Tdrd5 mutants compared to littermate controls in testes. The piRNAs were analyzed using publicly available datasets32. g, Metagene analysis of piRNA abundance from 50 nt upstream stop codon to 1,000 nt downstream stop codon. h, Schematic of affinity purification of germ-line specific RPFs. i, Length histograms of anti-HA immunoprecipitated RPFs from uORF (top) and UDR (bottom) pachytene piRNA precursors in testis. j, A254 absorbance profile of 15% to 60% sucrose density gradients of adult mouse testis lysed in conventional lysis conditions in which mitochondria were disrupted. Normal salt (blue) and high salt (grey). The substantial decrease in the monosome peak with high salt treatment confirms that the high salt buffer can dissociate translation-inactive ribosomes73. k, Aggregated data for RPF abundance on mRNAs (10% trimmed mean) (normal salt, upper, and high salt, lower). The high salt treatment releases the ribosomes at the stop codon of mRNAs as these terminated ribosomes have released their nascent polypeptides. Statistical Source Data are provided in Source Data Extended Data Figure 6. Unprocessed blots are provided in Source Data Extended Data Figure 6.