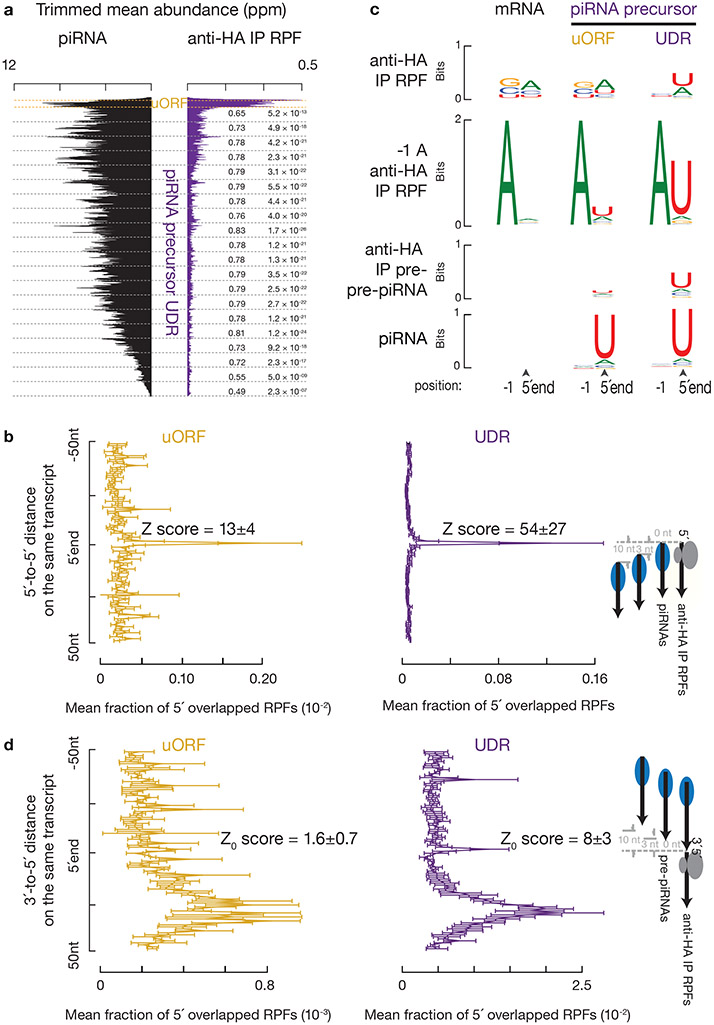
Fig. 5. Endonucleolytic cleavage targets ribosomes on piRNA precursors.
a, Aggregated data for piRNAs (left) and ribosome occupancy (right) on pachytene piRNA-producing transcripts (10% trimmed mean). UDRs of each transcript (sample size n = 100 pachytene piRNA precursor UDRs) were split to 20 non-overlapping windows, and the partial Spearman’s correlation efficient and the corresponding p-value based on two-sided Spearman’s correlation test for each window were listed. RPFs and piRNAs are correlated in all 20 bins controlling for piRNA precursor after Bonferroni adjustment for multiple comparisons. b, Distance spectrum of 5´-ends of anti-HA immunoprecipitated RPFs from uORFs (left) and from UDRs (right) that overlap piRNAs in adult testis (sample size n = 3 independent biological samples). Data are mean ± standard deviation. The Z scores indicate how many standard deviations an element is from the mean; Z-score > 3.3 corresponds to p < 0.01 according to a two-sided Z-test. c, Sequence logos depicting nucleotide bias at 5´-ends and 1 nt upstream of 5´-ends of the following species. Top to bottom: anti-HA immunoprecipitated RPF species, anti-HA immunoprecipitated RPF species with a 5´-upstream A, anti-HA immunoprecipitated 5´P RNA species from degradome sequencing, and piRNAs, which map to mRNAs and piRNA precursor uORFs and UDRs, respectively. d, Distance spectrum of 5´-ends of anti-HA immunoprecipitated RPFs from uORFs (left) and from UDRs (right) that overlap with the 3´-ends of pre-piRNAs in Pnldc1 mutant testis (sample size n = 3 independent biological samples)10. Data are mean ± standard deviation. Z-score > 3.3 corresponds to p < 0.01. See also Extended Data Fig. 7, 8. Statistical source data are provided in Source Data Fig.5.