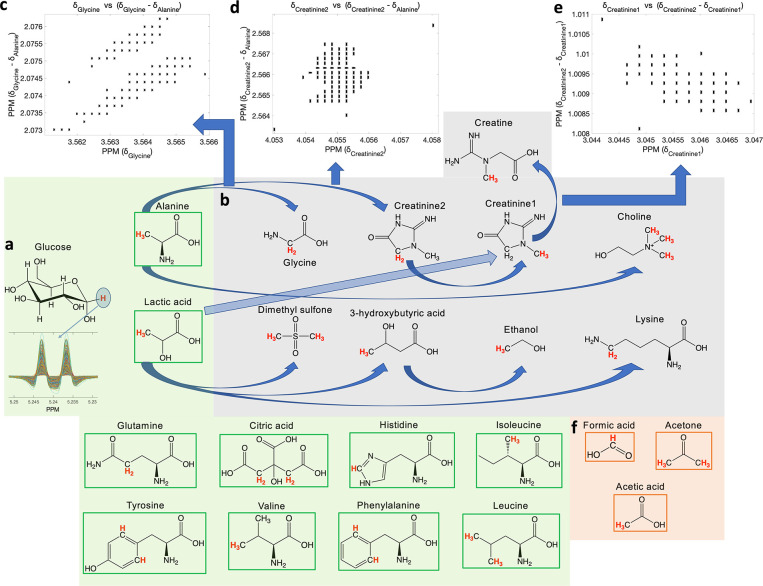
Figure 2.
Employed strategy for the automated assignment of 22 serum/plasma metabolites. (a) 1H NMR/SMolESY spectra are calibrated to the glucose anomeric proton doublet. Signals corresponding to the 1H highlighted in red font are used for assignment/quantitation of all metabolites. The glucose doublet and metabolites in the green boxes are assigned by pattern recognition (e.g., by imposing J-coupling constraints) in previously defined spectral windows with a width ≥0.01 ppm at 600 MHz. (b) Simple correlations, based upon 4023 plasma/serum unique spectra, with alanine and lactic acid methyl group signals are used as assignment constraints for metabolites in gray squares, which cannot otherwise be assigned using J-coupling constraints because they present either singlets or multiplets whose SMolESY components have a high risk of overlap (Table S2 and Figure S2). (c) Glycine singlet assignment is further supported by the minimization of the predefined spectral window owing to the decrease in line broadening achieved via SMolESY (≤0.004 ppm at 600 MHz). (d) Assignment of creatinine requires all previous constraints plus (e) extra correlations between intra molecular 1H NMR spin systems (e.g., between the −CH3 and −CH2 groups of creatinine). (f) The singlets from acetic acid, acetone, and formic acid were not found to significantly correlate with any other abundant metabolite; however, the predefined windows for these metabolites’ signals were sufficiently narrow (≤0.006 ppm for acetone/acetate and ≤0.008 ppm for formic acid) following spectral calibration to glucose which, combined with SMolESY and the general homeostatic nature of blood matrices, allows for their reliable identification.