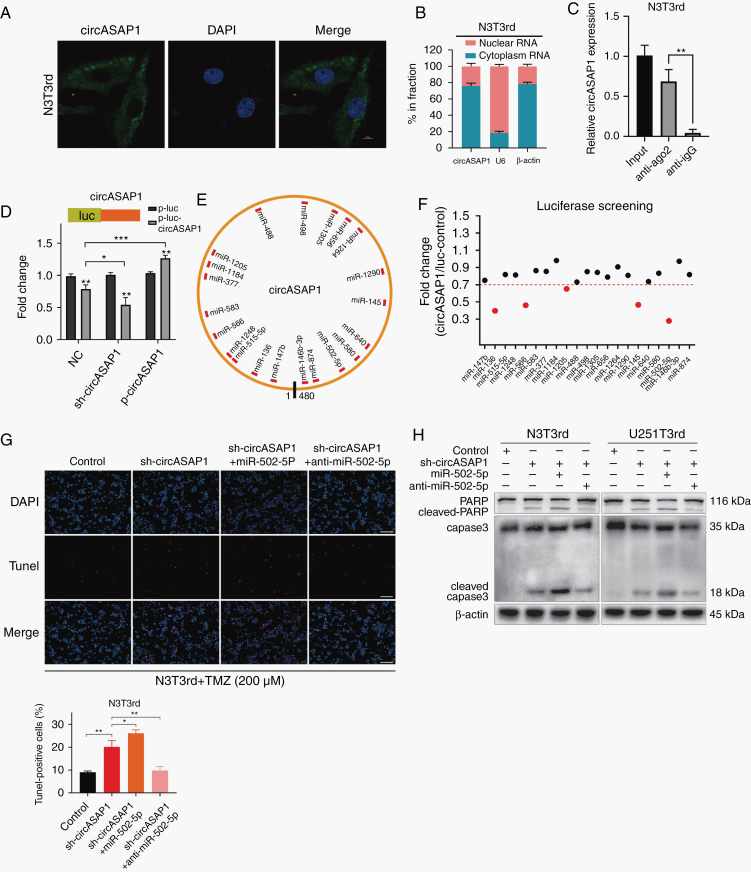
Fig. 4.
CircASAP1 serves as a sponge for miR-502-5p. (A) FISH analysis indicated the subcellular location of circASAP1 in N3T3rd cells (green). Nuclei were stained with DAPI (blue). Scale bar = 10 μm. (B) Relative circASAP1 expression levels in nuclear and cytosolic fractions of N3T3rd cells are shown. U6 was used as the nuclear control. β-actin was used as the cytosolic control. (C) RIP experiments were performed using an anti-Ago2 antibody, and specific primers were used to detect the enrichment of circASAP1 in N3T3rd cells by qRT-PCR. (D) A luciferase reporter assay was conducted to measure the luciferase activity of p-luc-circASAP1 in HEK-293T cells cotransfected with the sh-circASAP1 or pLCDH-circASAP1 vector. (E) A schematic drawing shows the putative binding sites for miRNAs associated with circASAP1. (F) A luciferase reporter assay was used to evaluate the luciferase activity of p-luc-circASAP1 in HEK-293T cells transfected with a library of 21 miRNA mimics to identify miRNAs that were able to bind to the circASAP1 sequence. The 5 miRNAs that inhibited luciferase activity by 30% are indicated by red dots. (G) TUNEL staining was used to analyze the effect of a miR-502-5p mimic or inhibitor on N3T3rd cells with circASAP1 knockdown after treatment with 200 μM TMZ for 48 h. Scale bar = 100 μm. (H) Cleaved caspase-3 and PARP in N3T3rd and U251T3rd cells after 200 μM TMZ treatment for 48 h. Data are presented as the mean ± SEM of 3 independent experiments. Significant results are presented as *P < 0.05, **P < 0.01, and ***P < 0.001.