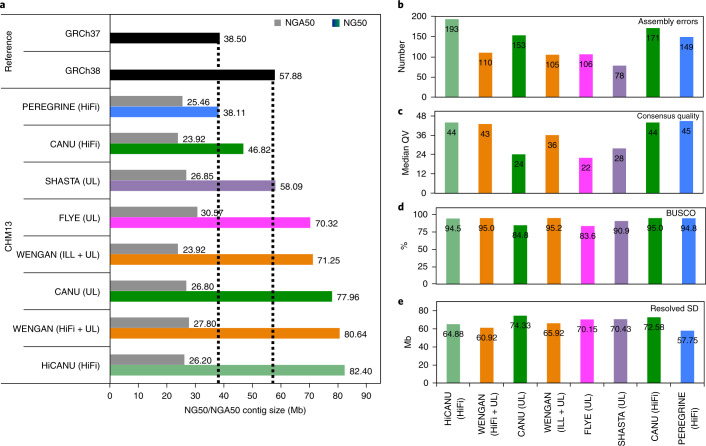
Fig. 2. WENGAN assemblies of the haploid CHM13 genome.
a, A bar plot showing the contig NG50/NGA50 of WENGAN and other state-of-the-art long-read assemblers, as well as of the current human reference genomes. NG50 is the contig length such that using longer contigs produces half (50%) of the bases of the reference genome. NGA50 is NG50 corrected of assembly errors. NG50 and NGA50 were computed using as genome size the total contig lengths of GRCh38 (2.94 Gb). b, Assembly errors predicted by QUAST using the GRCh38 reference (autosomes plus X and Y). Assembly errors overlapping centromeric regions or SDs were excluded from the analysis. c, Consensus quality assessment by alignment of 30 unique BAC sequences to the assembled contigs using the BACVALIDATION tool. d, Gene completeness was determined using the BUSCO tool. e, SDs resolved by the genome assemblies. An SD is considered resolved if the aligned contig extends the SD flanking sequences by at least 50 kb (see Methods). Different CHM13 assemblers are represented using the same color across a–e.