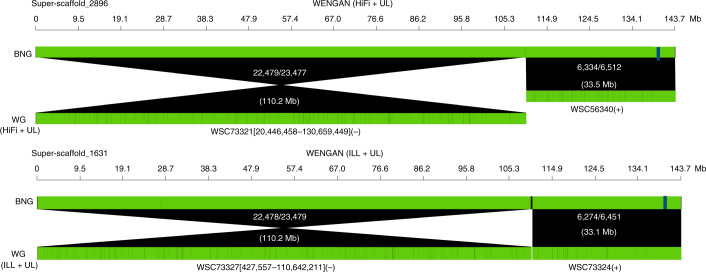
Fig. 3. BIONANO scaffolding of the WENGAN assemblies of CHM13.
We show the largest super-scaffold produced by merging the BIONANO map (BNG) and the WENGAN (WG) contigs generated by combining ultralong Nanopore reads (rel3) with PacBio/HiFi (20 kb) or Illumina (2 × 250 bp) reads. The name of the scaffolded WENGAN (WSC) contigs is displayed. The square brackets in the contig name indicate that the contig was corrected by the BIONANO map, and the numbers are the start–stop coordinates of the error-free contig region. In round brackets, we show the contig orientation in the super-scaffold. The white text in the alignments displays the number of matching nicking sites, the total number of nicking sites in the BNG contig and the length in megabases of the alignment. The blue bar in the BNG contigs shows examples of joins guided by the WENGAN contigs.