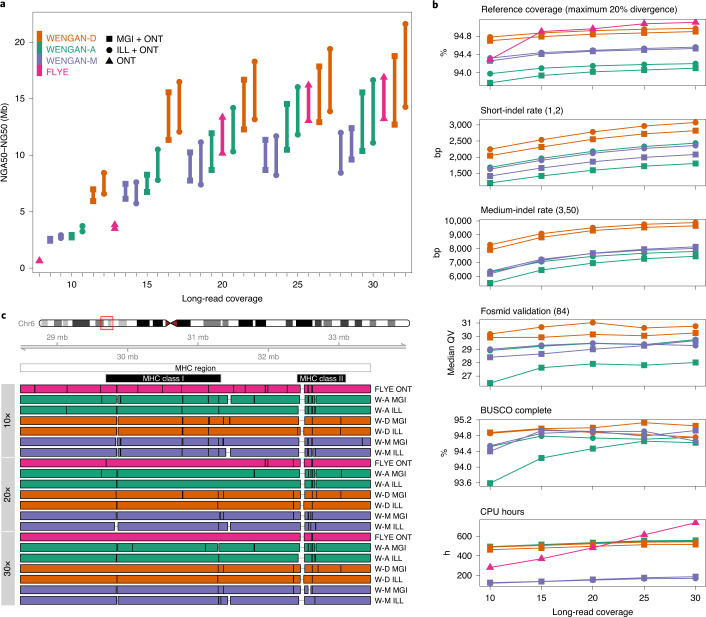
Fig. 4. De novo genome assemblies of NA12878 when varying the long-read coverage and the short-read technology.
a, The de novo assemblies were sorted by NG50 at each long-read coverage (lolliplot). We computed the NGA50 (which corresponds to the NG50 corrected of assembly errors) of each assembly using QUAST (see Methods). b, The consensus quality (see Methods) of each genome assembly and the CPU hours required for the assembly. c, The WENGAN (W-X) and FLYE assemblies of the complex MHC region located in Chr6: 28,477,797–33,448,354 (4.97 Mb). The MHC sequence was aligned to the genome assemblies and the aligned blocks ≥30 kb with a minimum identity of 95% were kept. The alignment breakpoints (vertical black lines) indicate a contig switch, an alignment error or a gap in the assembly. The assemblies of the MHC region are displayed in tracks by long-read coverage.